7F4D
 
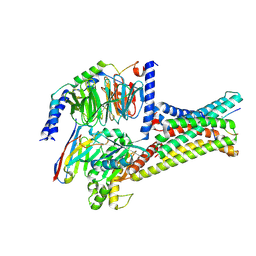 | | Cryo-EM structure of alpha-MSH-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4F
 
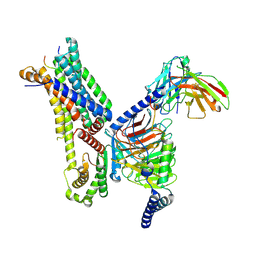 | | Cryo-EM structure of afamelanotide-bound melanocortin-1 receptor in complex with Gs protein and scFv16 | | Descriptor: | Afamelanotide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7XH4
 
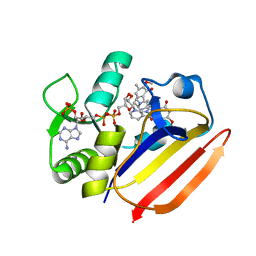 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis complex with safracin A | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Safracin A, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Zhang, Y, Yang, D, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
2H17
 
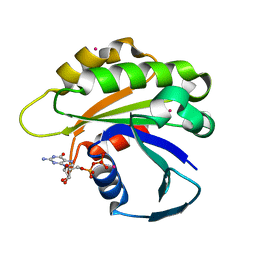 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2H18
 
 | | Structure of human ADP-ribosylation factor-like 10B (ARL10B) | | Descriptor: | ADP-ribosylation factor-like protein 8A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Atanassova, A, Tempel, W, Dimov, S, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 10B (ARL10B)
To be Published
|
|
7N7V
 
 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
7Q02
 
 | | Zn-free structure of lipocalin-like Milk protein, inspired from Diploptera punctata, expressed in Saccharomyces cerevisiae | | Descriptor: | Milk protein, PALMITOLEIC ACID | | Authors: | Banerjee, S, Dhanabalan, K.V, Ramaswamy, S. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of recombinantly expressed cockroach Lili-Mip protein in glycosylated and deglycosylated forms.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
5ZF2
 
 | | Crystal structure of Trxlp from Edwardsiella tarda EIB202 | | Descriptor: | SULFATE ION, Thioredoxin (H-type,TRX-H) | | Authors: | Yang, C, Quan, S. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Edwardsiella piscicida thioredoxin-like protein inhibits ASK1-MAPKs signaling cascades to promote pathogenesis during infection.
Plos Pathog., 15, 2019
|
|
6DHX
 
 | |
5DV2
 
 | |
5VRG
 
 | |
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
5H9X
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | Descriptor: | beta-1,3-glucanase | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5H9Y
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | Descriptor: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5UMY
 
 | | Crystal structure of TnmS3 in complex with tiancimycin | | Descriptor: | (1aS,11S,11aR,14Z,18R)-3,8,18-trihydroxy-11a-[(1R)-1-hydroxyethyl]-7-methoxy-11,11a-dihydro-4H-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinoline-4,9(10H)-dione, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMQ
 
 | | Crystal structure of TnmS1, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMX
 
 | | Crystal structure of TnmS3 in complex with riboflavin | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMP
 
 | | Crystal structure of TnmS3, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMW
 
 | | Crystal structure of TnmS2, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
6CLW
 
 | | Crystal structure of TnmH | | Descriptor: | O-methyltransferase | | Authors: | Chang, C.Y, Annaval, T, Adhikari, A, Yan, X, Shen, B. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Characterization of TnmH as anO-Methyltransferase Revealing Insights into Tiancimycin Biosynthesis and Enabling a Biocatalytic Strategy To Prepare Antibody-Tiancimycin Conjugates.
J.Med.Chem., 63, 2020
|
|
6CLX
 
 | | Crystal structure of TnmH in complex with SAM | | Descriptor: | O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C.Y, Annaval, T, Adhikari, A, Yan, X, Shen, B. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Characterization of TnmH as anO-Methyltransferase Revealing Insights into Tiancimycin Biosynthesis and Enabling a Biocatalytic Strategy To Prepare Antibody-Tiancimycin Conjugates.
J.Med.Chem., 63, 2020
|
|
5DV4
 
 | |
7VQX
 
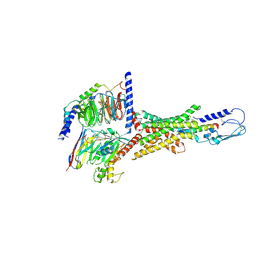 | | Cryo-EM structure of human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-10-21 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
