4XLV
 
 | |
7R7S
 
 | | p47-bound p97-R155H mutant with ATPgammaS | | Descriptor: | NSFL1 cofactor p47, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7T
 
 | | p47-bound p97-R155H mutant with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NSFL1 cofactor p47, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7U
 
 | | D1 and D2 domain structure of the p97(R155H)-p47 complex | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
4YTL
 
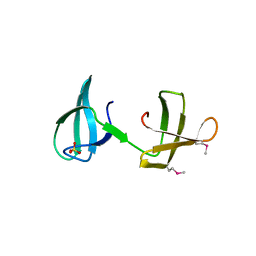 | | Structure of the KOW2-KOW3 Domain of Transcription Elongation Factor Spt5. | | Descriptor: | GLYCEROL, SULFATE ION, Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, ZHang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
3OQT
 
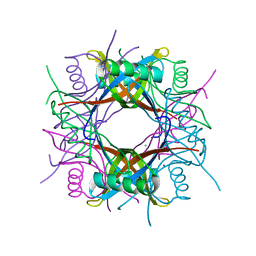 | | Crystal structure of Rv1498A protein from mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Rv1498A PROTEIN, SODIUM ION | | Authors: | Liu, F, Xiong, J, Kumar, S, Yang, C, Li, S, Ge, S, Xia, N, Swaminathan, K. | | Deposit date: | 2010-09-04 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A.
J.Struct.Biol., 175, 2011
|
|
7YTW
 
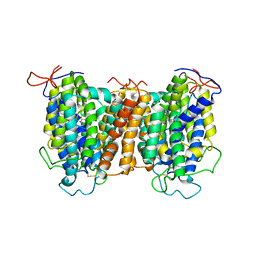 | | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter | | Descriptor: | ASCORBIC ACID, SODIUM ION, Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
7YTY
 
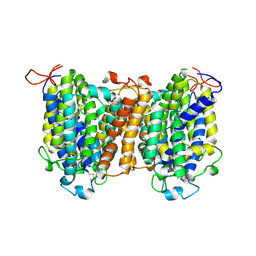 | | Mouse SVCT1 in an apo state | | Descriptor: | Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
6F2Z
 
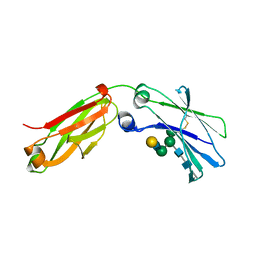 | | Structure of a Fc mutant | | Descriptor: | Fc fragment of antibody, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhou, A, Li, S. | | Deposit date: | 2017-11-27 | | Release date: | 2018-12-12 | | Last modified: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Fc mutant in monomeric form
To Be Published
|
|
7UEB
 
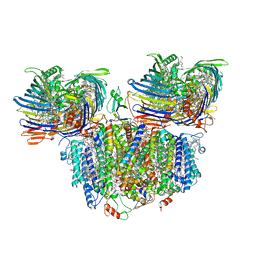 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO2) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
7UEA
 
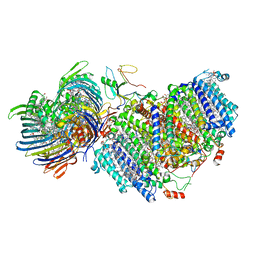 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
1Z8D
 
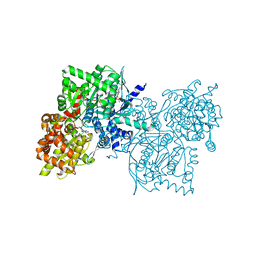 | | Crystal Structure of Human Muscle Glycogen Phosphorylase a with AMP and Glucose | | Descriptor: | ADENINE, ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, ... | | Authors: | Lukacs, C.M, Oikonomakos, N.G, Crowther, R.L, Hong, L.N, Kammlott, R.U, Levin, W, Li, S, Liu, C.M, Lucas-McGady, D, Pietranico, S, Reik, L. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human muscle glycogen phosphorylase a with bound glucose and AMP: An intermediate conformation with T-state and R-state features.
Proteins, 63, 2006
|
|
5DJ5
 
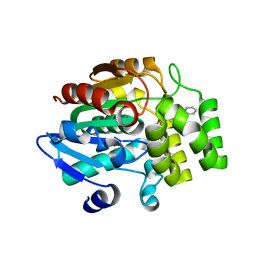 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
1ZUC
 
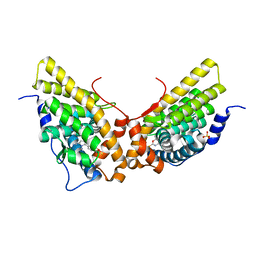 | | Progesterone receptor ligand binding domain in complex with the nonsteroidal agonist tanaproget | | Descriptor: | 5-(4,4-DIMETHYL-2-THIOXO-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-6-YL)-1-METHYL-1H-PYRROLE-2-CARBONITRILE, Progesterone receptor, SULFATE ION | | Authors: | Zhang, Z, Olland, A.M, Zhu, Y, Cohen, J, Berrodin, T, Chippari, S, Appavu, C, Li, S, Wilhem, J, Chopra, R, Fensome, A, Zhang, P, Wrobel, J, Unwalla, R.J, Lyttle, C.R, Winneker, R.C. | | Deposit date: | 2005-05-30 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and pharmacological properties of a potent and selective novel nonsteroidal progesterone receptor agonist tanaproget
J.Biol.Chem., 280, 2005
|
|
4R7A
 
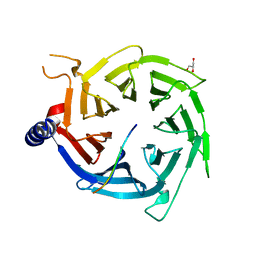 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
5WSO
 
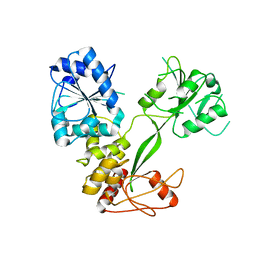 | |
4HPH
 
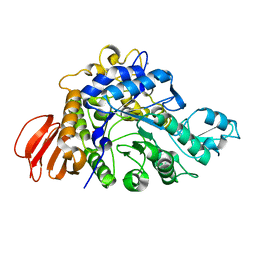 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOW
 
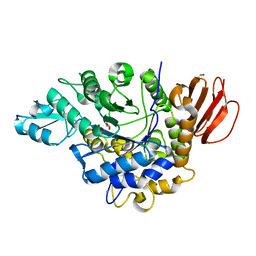 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOZ
 
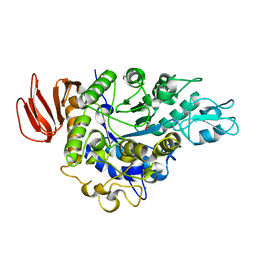 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4QLA
 
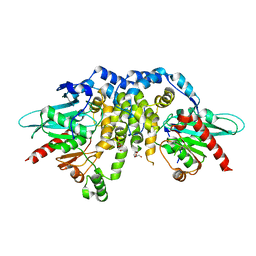 | | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori | | Descriptor: | Juvenile hormone epoxide hydrolase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, K, Jia, N, Hu, C, Jiang, Y.L, Yang, J.P, Chen, Y, Li, S, Zhou, C.Z. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori.
Proteins, 82, 2014
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-2 RNA SL5 domain. | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
