2VZ5
 
 | | Structure of the PDZ domain of Tax1 (human T-cell leukemia virus type I) binding protein 3 | | Descriptor: | CHLORIDE ION, IMIDAZOLE, TAX1-BINDING PROTEIN 3, ... | | Authors: | Murray, J.W, Shafqat, N, Yue, W, Pilka, E, Johannsson, C, Salah, E, Cooper, C, Elkins, J.M, Pike, A.C, Roos, A, Filippakopoulos, P, von Delft, F, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Oppermann, U. | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | The Structure of the Pdz Domain of Tax1BP
To be Published
|
|
2VX2
 
 | | Crystal structure of human enoyl Coenzyme A hydratase domain- containing protein 3 (ECHDC3) | | Descriptor: | ENOYL-COA HYDRATASE DOMAIN-CONTAINING PROTEIN 3 | | Authors: | Yue, W.W, Guo, K, Kochan, G, Pilka, E, Murray, J.W, Salah, E, Cocking, R, Sun, Z, Roos, A.K, Pike, A.C.W, Filippakopoulos, P, Arrowsmith, C, Wikstrom, M, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Enoyl Coenzyme a Hydratase Domain-Containing Protein 3 (Echdc3)
To be Published
|
|
2WU7
 
 | | Crystal Structure of the Human CLK3 in complex with V25 | | Descriptor: | CHLORIDE ION, DUAL SPECIFICITY PROTEIN KINASE CLK3, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Phillips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Bracher, F, Huber, K, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
2WM8
 
 | | Crystal structure of human magnesium-dependent phosphatase 1 of the haloacid dehalogenase superfamily (MGC5987) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM-DEPENDENT PHOSPHATASE 1 | | Authors: | Yue, W.W, Shafqat, N, Pike, A.C.W, Chaikuad, A, Bray, J.E, Pilka, E.W, Burgess-Brown, N, Hapka, E, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Human Magnesium-Dependent Phosphatase 1 of the Haloacid Dehalogenase Superfamily (Mgc5987)
To be Published
|
|
2VPK
 
 | | Crystal structure of the BTB domain of human myoneurin | | Descriptor: | MYONEURIN | | Authors: | Cooper, C.D.O, Murray, J.W, Bullock, A, Pike, A.C.W, von Delft, F, Filippakopoulos, P, Salah, E, Edwards, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Knapp, S. | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Myoneurin
To be Published
|
|
2VPH
 
 | | Crystal structure of the human protein tyrosine phosphatase, non- receptor type 4, PDZ domain | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 4 | | Authors: | Roos, A.K, Wang, J, Burgess-Brown, N, Elkins, J.M, Kavanagh, K, Pike, A.C.W, Filippakopoulos, P, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Bountra, C, Knapp, S. | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Protein Tyrosine Phosphatase, Non-Receptor Type 4, Pdz Domain
To be Published
|
|
2WU6
 
 | | Crystal Structure of the Human CLK3 in complex with DKI | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CHLORIDE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Philips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
2Y1H
 
 | | Crystal structure of the human TatD-domain protein 3 (TATDN3) | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Muniz, J.R.C, Puranik, S, Krojer, T, Yue, W.W, Ugochukwu, E, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-12-08 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Human Tatd-Domain Protein 3 (Tatdn3)
To be Published
|
|
2V24
 
 | | Structure of the human SPRY domain-containing SOCS box protein SSB-4 | | Descriptor: | NICKEL (II) ION, SPRY DOMAIN-CONTAINING SOCS BOX PROTEIN 4 | | Authors: | Uppenberg, J, Bullock, A, Keates, T, Savitsky, P, Pike, A.C.W, Ugochukwu, E, Bunkoczi, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-05-31 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Par-4 Recognition by the Spry Domain-and Socs Box-Containing Proteins Spsb1, Spsb2, and Spsb4.
J.Mol.Biol., 401, 2010
|
|
5TB6
 
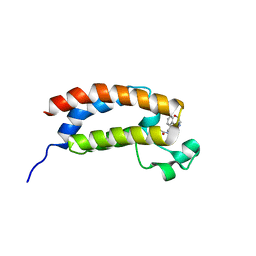 | | Structure of bromodomain of CREBBP with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5LUU
 
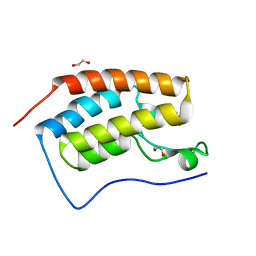 | | Structure of the first bromodomain of BRD4 with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
2I75
 
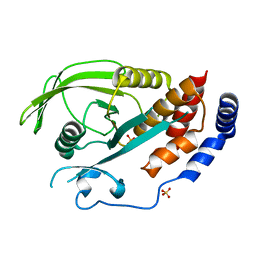 | | Crystal Structure of Human Protein Tyrosine Phosphatase N4 (PTPN4) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Ugochukwu, E, Barr, A, Savitsky, P, Burgess, N, Das, S, Turnbull, A, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
6TTK
 
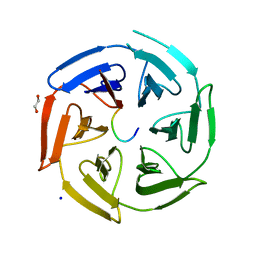 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
7OG0
 
 | | Nontypeable Haemophillus influenzae SapA in open and closed conformations, in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFZ
 
 | | Nontypeable Haemophillus influenzae SapA in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFW
 
 | | Nontypeable Haemophillus influenzae SapA in complex with heme | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
2VAG
 
 | | Crystal structure of di-phosphorylated human CLK1 in complex with a novel substituted indole inhibitor | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE CLK1, ethyl 3-[(E)-2-amino-1-cyanoethenyl]-6,7-dichloro-1-methyl-1H-indole-2-carboxylate | | Authors: | Pike, A.C.W, Bullock, A.N, Fedorov, O, Pilka, E.S, Ugochukwu, E, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Huber, K, Bracher, F, Knapp, S. | | Deposit date: | 2007-08-31 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
6BW3
 
 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
6BW4
 
 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
5TWH
 
 | |
5TWG
 
 | |
7AK3
 
 | | CLK1 bound with CAF052 | | Descriptor: | Dual specificity protein kinase CLK1, ~{N}-[3-fluoranyl-4-(4-methylpiperazin-1-yl)phenyl]-4-pyrazolo[1,5-b]pyridazin-3-yl-pyrimidin-2-amine | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Inhibitor Identifications Reveal Targeting Opportunity for the Atypical MAPK Kinase ERK3.
Int J Mol Sci, 21, 2020
|
|
2GJT
 
 | | Crystal structure of the human receptor phosphatase PTPRO | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase PTPRO | | Authors: | Barr, A, Ugochukwu, E, Eswaran, J, Das, S, Niesen, F, Savitsky, P, Turnbull, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, von Delft, F, Papagrigoriou, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2H4V
 
 | | Crystal Structure of the Human Tyrosine Receptor Phosphatase Gamma | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Das, S, Eswaran, J, Savitsky, P, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Debreczeni, J, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2CFV
 
 | | Crystal structure of human protein tyrosine phosphatase receptor type J | | Descriptor: | CHLORIDE ION, HUMAN PROTEIN TYROSINE PHOSPHATASE RECEPTOR TYPE J, NICKEL (II) ION | | Authors: | Debreczeni, J.E, Barr, A.J, Eswaran, J, Ugochukwu, E, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2006-02-23 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
