4F7Z
 
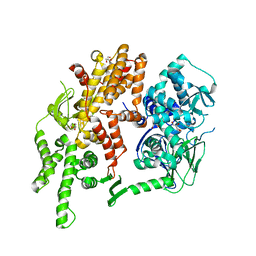 | | Conformational dynamics of exchange protein directly activated by cAMP | | Descriptor: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | Authors: | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
1I5Z
 
 | | STRUCTURE OF CRP-CAMP AT 1.9 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | White, M.A, Lee, J.C, Fox, R.O. | | Deposit date: | 2001-03-01 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
1I6X
 
 | | STRUCTURE OF A STAR MUTANT CRP-CAMP AT 2.2 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | White, M.A, Lee, J.C, Fox, R.O. | | Deposit date: | 2001-03-06 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
5JLW
 
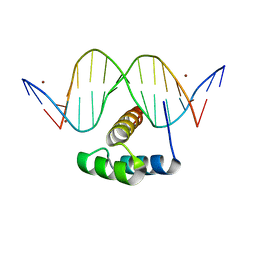 | | AntpHD with 15bp DNA duplex R-monothioated at Cytidine-8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C7R)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J, Nguyen, D. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Stereospecific Effects of Oxygen-to-Sulfur Substitution in DNA Phosphate on Ion Pair Dynamics and Protein-DNA Affinity.
Chembiochem, 17, 2016
|
|
5JLX
 
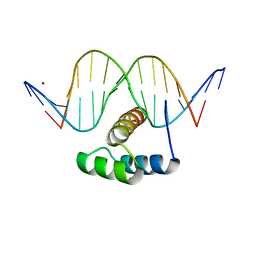 | | AntpHD with 15bp DNA duplex S-monothioated at Cytidine-8 | | Descriptor: | DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C7S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), Homeotic protein antennapedia, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J, Nguyen, D. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | Stereospecific Effects of Oxygen-to-Sulfur Substitution in DNA Phosphate on Ion Pair Dynamics and Protein-DNA Affinity.
Chembiochem, 17, 2016
|
|
7JQQ
 
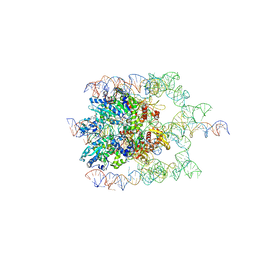 | | The bacteriophage Phi-29 viral genome packaging motor assembly | | Descriptor: | DNA (60-MER), DNA packaging protein, MAGNESIUM ION, ... | | Authors: | White, M.A, Woodson, M, Morais, M.C. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA.
Sci Adv, 7, 2021
|
|
2Q9F
 
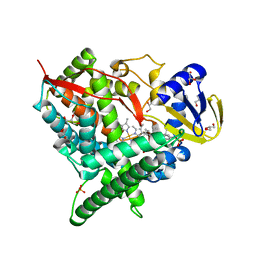 | | Crystal structure of human cytochrome P450 46A1 in complex with cholesterol-3-sulphate | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Cytochrome P450 46A1, GLYCEROL, ... | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Q9G
 
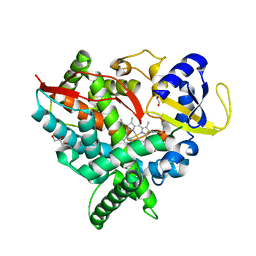 | | Crystal structure of human cytochrome P450 46A1 | | Descriptor: | Cytochrome P450 46A1, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4X9J
 
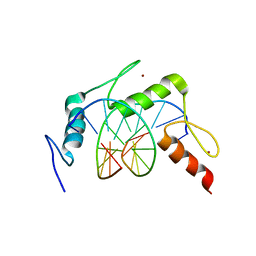 | | EGR-1 with Doubly Methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*(5CM)P*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*(5CM)P*GP*C)-3'), Early growth response protein 1, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1.
Febs Lett., 589, 2015
|
|
4XIC
 
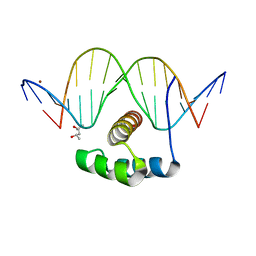 | | ANTPHD WITH 15BP di-thioate modified DNA DUPLEX | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C2S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
4XID
 
 | | AntpHD with 15bp DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
1KH8
 
 | | Structure of a cis-proline (P114) to glycine variant of Ribonuclease A | | Descriptor: | CESIUM ION, SULFATE ION, pancreatic ribonuclease A | | Authors: | Schultz, D.A, Friedman, A.M, White, M.A, Fox, R.O. | | Deposit date: | 2001-11-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the cis-proline to glycine variant (P114G) of ribonuclease A.
Protein Sci., 14, 2005
|
|
4DYD
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4E12
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4E13
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
3ROU
 
 | |
3RPQ
 
 | |
3QOP
 
 | |
3RDI
 
 | |
3RYP
 
 | |
3RYR
 
 | |
2BDM
 
 | | Structure of Cytochrome P450 2B4 with Bound Bifonazole | | Descriptor: | 1-[PHENYL-(4-PHENYLPHENYL)-METHYL]IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Zhao, Y, White, M.A, Muralidhara, B.K, Sun, L, Halpert, J.R, Stout, C.D. | | Deposit date: | 2005-10-20 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction.
J.Biol.Chem., 281, 2006
|
|
7JQ7
 
 | | The Phi-28 gp11 DNA packaging Motor | | Descriptor: | Encapsidation protein, IODIDE ION, SULFATE ION | | Authors: | Morais, M.C, White, M.A, Dill, E. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Atomistic basis of force generation, translocation, and coordination in a viral genome packaging motor.
Nucleic Acids Res., 49, 2021
|
|
7JQP
 
 | | The Phi-28 gp11 DNA packaging Motor | | Descriptor: | Encapsidation protein, SULFATE ION | | Authors: | Morais, M.C, White, M.A, Dill, E. | | Deposit date: | 2020-08-11 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Atomistic basis of force generation, translocation, and coordination in a viral genome packaging motor.
Nucleic Acids Res., 49, 2021
|
|
7JQ6
 
 | | The Phi-28 gp11 DNA packaging Motor | | Descriptor: | Encapsidation protein, SULFATE ION | | Authors: | Morais, M.C, White, M.A, Dill, E. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atomistic basis of force generation, translocation, and coordination in a viral genome packaging motor.
Nucleic Acids Res., 49, 2021
|
|
