4WIG
 
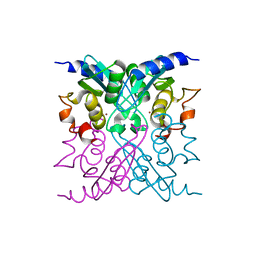 | | Crystal structure of E47D mutant cytidine deaminase from Mycobacterium tuberculosis (MtCDA E47D) | | Descriptor: | Cytidine deaminase, ZINC ION | | Authors: | Trivella, D.B.B, Sanchez-Quitian, A.Z, Basso, L.A, Santos, D.S. | | Deposit date: | 2014-09-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Functional and structural evidence for the catalytic role played by glutamate-47 residue in the mode of action of Mycobacterium tuberculosis cytidine deaminase
Rsc Adv, 5, 2015
|
|
8FWN
 
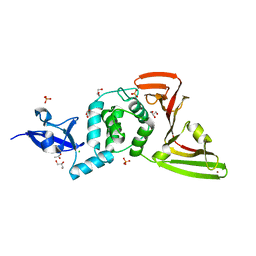 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|
8FWO
 
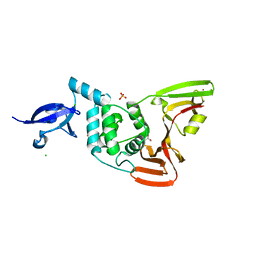 | | Crystal structure of SARS-CoV-2 papain-like protease | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease
To Be Published
|
|
7UXX
 
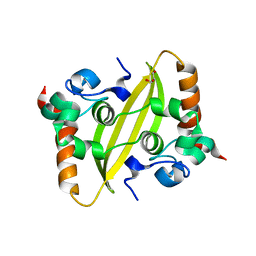 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7UXZ
 
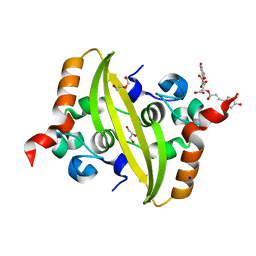 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7S2X
 
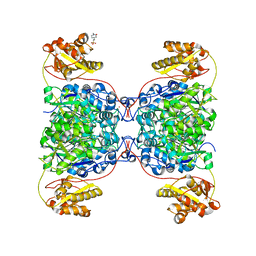 | | Structure of SalC, a gamma-lactam-beta-lactone bicyclase for salinosporamide biosynthesis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, POTASSIUM ION, SalC | | Authors: | Chen, P.Y.-T, Trivella, D.B.B, Bauman, K.D, Moore, B.S. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Enzymatic assembly of the salinosporamide gamma-lactam-beta-lactone anticancer warhead.
Nat.Chem.Biol., 18, 2022
|
|
7JR4
 
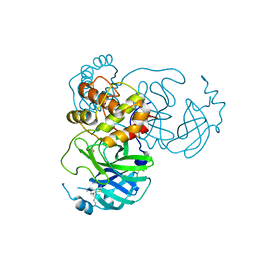 | |
7KO8
 
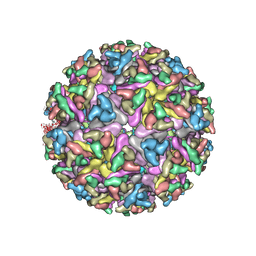 | | Cryo-EM structure of the mature and infective Mayaro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Riberio-Filho, H.V, Coimbra, L.D, Cassago, A, Rocha, R.P.F, Padilha, A.C.M, Schatz, M, van Heel, M.G, Portugal, R.V, Trivella, D.B.B, de Oliveira, P.S.L, Marques, R.E. | | Deposit date: | 2020-11-06 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the mature and infective Mayaro virus at 4.4 angstrom resolution reveals features of arthritogenic alphaviruses.
Nat Commun, 12, 2021
|
|
7JR3
 
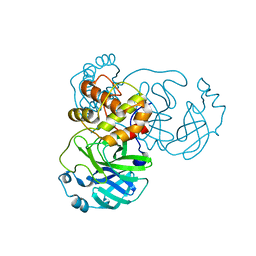 | |
3TKM
 
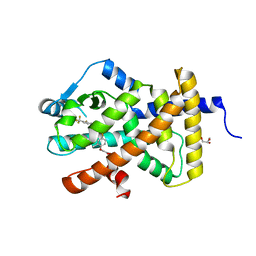 | | Crystal structure PPAR delta binding GW0742 | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor delta, {4-[({2-[3-fluoro-4-(trifluoromethyl)phenyl]-4-methyl-1,3-thiazol-5-yl}methyl)sulfanyl]-2-methylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Batista, F.H, Polikarpov, I. | | Deposit date: | 2011-08-27 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural Insights into Human Peroxisome Proliferator Activated Receptor Delta (PPAR-Delta) Selective Ligand Binding.
Plos One, 7, 2012
|
|
4Z9A
 
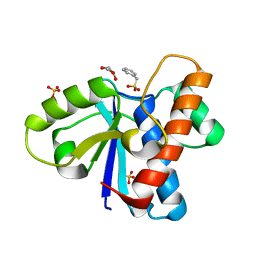 | | Crystal structure of Low Molecular Weight Protein Tyrosine Phosphatase isoform A complexed with phenylmethanesulfonic acid | | Descriptor: | GLYCEROL, Low molecular weight phosphotyrosine protein phosphatase, SULFATE ION, ... | | Authors: | Trivella, D.B.B, Fonseca, E.M.B, Scorsato, V, Dias, M.P, Aparicio, R. | | Deposit date: | 2015-04-10 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the apo form and a complex of human LMW-PTP with a phosphonic acid provide new evidence of a secondary site potentially related to the anchorage of natural substrates.
Bioorg.Med.Chem., 23, 2015
|
|
3NEX
 
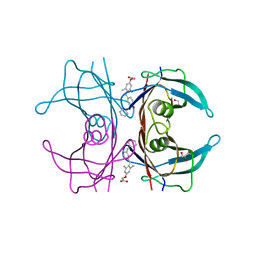 | |
3NEO
 
 | |
3NEE
 
 | | Wild type human transthyretin (TTR) complexed with GC-1 (TTRwt:GC-1) | | Descriptor: | GLYCEROL, Transthyretin, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Polikarpov, I. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The binding of synthetic triiodo l-thyronine analogs to human transthyretin: molecular basis of cooperative and non-cooperative ligand recognition.
J.Struct.Biol., 173, 2011
|
|
3NES
 
 | | V30M mutant human transthyretin (TTR) complexed with GC-1 (V30M:GC-1) | | Descriptor: | GLYCEROL, Transthyretin, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Polikarpov, I. | | Deposit date: | 2010-06-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The binding of synthetic triiodo l-thyronine analogs to human transthyretin: molecular basis of cooperative and non-cooperative ligand recognition.
J.Struct.Biol., 173, 2011
|
|
4HNP
 
 | | Crystal structure of yeast 20S proteasome in complex with vinylketone carmaphycin analogue VNK1 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylhept-1-en-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylheptan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Proteasome component C1, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-20 | | Release date: | 2014-01-29 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4HRD
 
 | | Crystal structure of yeast 20S proteasome in complex with the natural product carmaphycin A | | Descriptor: | N-[(2S)-1-({(2S)-1-{[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]amino}-4-[(R)-methylsulfinyl]-1-oxobutan-2-yl}amino)-3-methyl-1-oxobutan-2-yl]hexanamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-27 | | Release date: | 2014-01-29 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4HRC
 
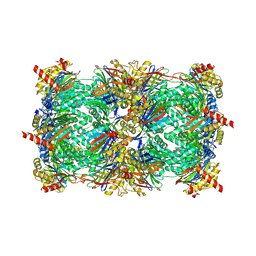 | | Crystal structure of yeast 20S proteasome in complex with epoxyketone carmaphycin analogue 3 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-27 | | Release date: | 2014-01-29 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4DEU
 
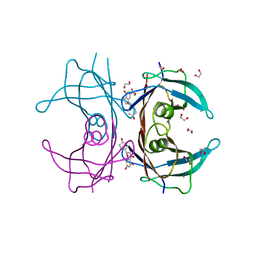 | |
4DER
 
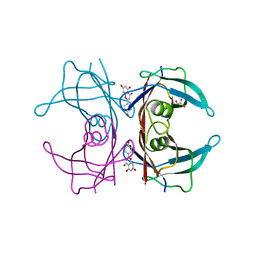 | |
4DEW
 
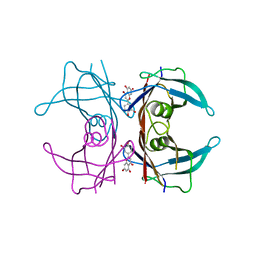 | |
4DET
 
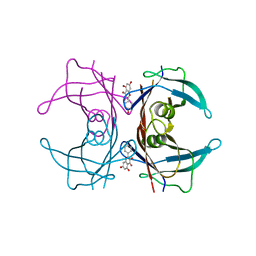 | |
4DES
 
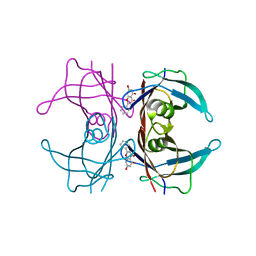 | |
4LTC
 
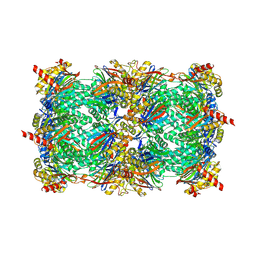 | | Crystal structure of yeast 20S proteasome in complex with enone carmaphycin analogue 6 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(4S,5S,6R)-5-hydroxy-2,6-dimethyloctan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Stein, M, Trivella, D.B.B, Groll, M. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4WIF
 
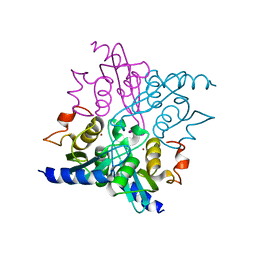 | |
