1BOB
 
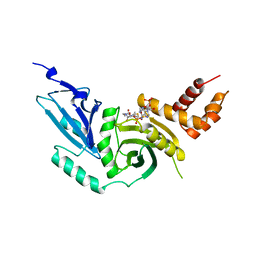 | | HISTONE ACETYLTRANSFERASE HAT1 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, HISTONE ACETYLTRANSFERASE | | Authors: | Dutnall, R.N, Tafrov, S.T, Sternglanz, R, Ramakrishnan, V. | | Deposit date: | 1998-07-02 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily.
Cell(Cambridge,Mass.), 94, 1998
|
|
1ICI
 
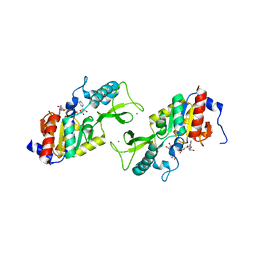 | | CRYSTAL STRUCTURE OF A SIR2 HOMOLOG-NAD COMPLEX | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRANSCRIPTIONAL REGULATORY PROTEIN, SIR2 FAMILY, ... | | Authors: | Min, J, Landry, J, Sternglanz, R, Xu, R.-M. | | Deposit date: | 2001-04-01 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a SIR2 homolog-NAD complex.
Cell(Cambridge,Mass.), 105, 2001
|
|
4IAO
 
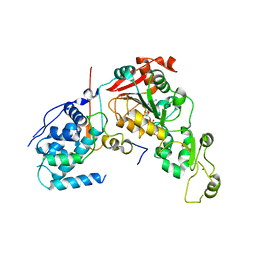 | | Crystal structure of Sir2 C543S mutant in complex with SID domain of Sir4 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent histone deacetylase SIR2, Regulatory protein SIR4, ... | | Authors: | Hsu, H.C, Wang, C.L, Wang, M, Yang, N, Chen, Z, Sternglanz, R, Xu, R.M. | | Deposit date: | 2012-12-07 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding
Genes Dev., 27, 2013
|
|
1QSM
 
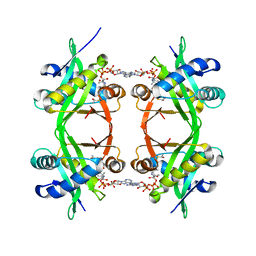 | | Histone Acetyltransferase HPA2 from Saccharomyces Cerevisiae in Complex with Acetyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, HPA2 HISTONE ACETYLTRANSFERASE | | Authors: | Angus-Hill, M.L, Dutnall, R.N, Tafrov, S.T, Sternglanz, R, Ramakrishnan, V. | | Deposit date: | 1999-06-22 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily.
J.Mol.Biol., 294, 1999
|
|
2FVU
 
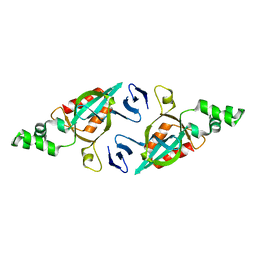 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
2NYR
 
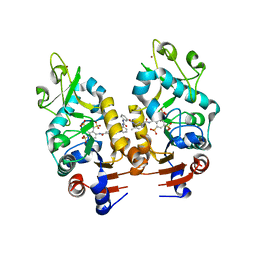 | | Crystal Structure of Human Sirtuin Homolog 5 in Complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, NAD-dependent deacetylase sirtuin-5, ZINC ION | | Authors: | Min, J.R, Antoshenko, T, Allali-Hassani, A, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin.
Structure, 15, 2007
|
|
3TOA
 
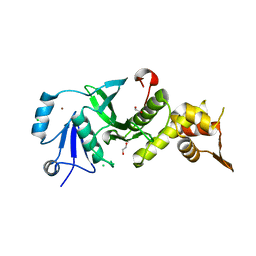 | | Human MOF crystal structure with active site lysine partially acetylated | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3TO6
 
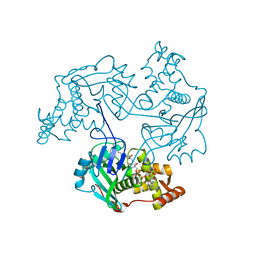 | |
3TOB
 
 | |
3TO7
 
 | | Crystal structure of yeast Esa1 HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | CACODYLIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3TO9
 
 | | Crystal structure of yeast Esa1 E338Q HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | 1,2-ETHANEDIOL, CACODYLIC ACID, COENZYME A, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
1QSO
 
 | | Histone Acetyltransferase HPA2 from Saccharomyces Cerevisiae | | Descriptor: | HPA2 HISTONE ACETYLTRANSFERASE | | Authors: | Angus-Hill, M.L, Dutnall, R.N, Tafrov, S.T, Sterngalnz, R, Ramakrishnan, V. | | Deposit date: | 1999-06-22 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily.
J.Mol.Biol., 294, 1999
|
|
3BTV
 
 | |
3BTU
 
 | |
3BTS
 
 | |
1RSG
 
 | |
