3BDN
 
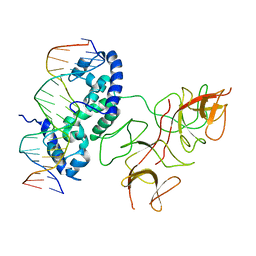 | | Crystal Structure of the Lambda Repressor | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DAP*DCP*DCP*DAP*DCP*DTP*DGP*DGP*DCP*DGP*DGP*DTP*DGP*DAP*DTP*DAP*DT)-3'), DNA (5'-D(*DTP*DAP*DTP*DAP*DTP*DCP*DAP*DCP*DCP*DGP*DCP*DCP*DAP*DGP*DTP*DGP*DGP*DTP*DAP*DT)-3'), Lambda Repressor | | Authors: | Stayrook, S.E, Jaru-Ampornpan, P, Hochschild, A, Lewis, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.909 Å) | | Cite: | Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding
Nature, 452, 2008
|
|
8UKX
 
 | |
8UKV
 
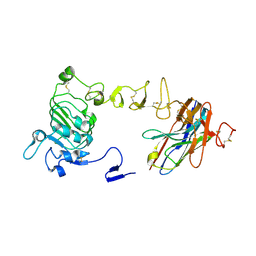 | | Crystal structure of nanobody/VHH domain of 34E5 in complex with the extracellular region of the epidermal growth factor variant III (EGFRvIII) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Stayrook, S.E, Ferguson, K.M, Bagchi, A. | | Deposit date: | 2023-10-15 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into the role and targeting of EGFRvIII.
Structure, 32, 2024
|
|
8UKW
 
 | |
5VJS
 
 | | De Novo Photosynthetic Reaction Center Protein Equipped with Heme B, a synthetic Zn porphyrin, and Zn(II) cations | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette, ... | | Authors: | Ennist, N.M, Dutton, P.L, Stayrook, S.E, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
8D9O
 
 | | De Novo Photosynthetic Reaction Center Protein in Apo-State | | Descriptor: | CADMIUM ION, Reaction Center Maquette | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
8D9P
 
 | | De Novo Photosynthetic Reaction Center Protein Equipped with Heme B and Mn(II) cations | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
7TVD
 
 | |
5VJU
 
 | | De Novo Photosynthetic Reaction Center Protein Variant Equipped with His-Tyr H-bond, Heme B, and Cd(II) ions | | Descriptor: | CADMIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette Leu71His variant | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
5VJT
 
 | | De Novo Photosynthetic Reaction Center Protein Equipped with Heme B and Zn(II) cations | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette, ... | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
6VG3
 
 | |
3C9A
 
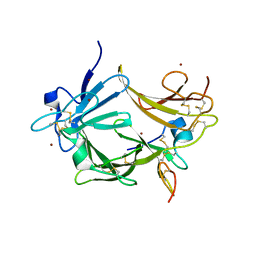 | | High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz | | Descriptor: | BROMIDE ION, Protein giant-lens, Protein spitz | | Authors: | Klein, D.E, Stayrook, S.E, Shi, F, Narayan, K, Lemmon, M.A. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for EGFR ligand sequestration by Argos.
Nature, 453, 2008
|
|
3CA7
 
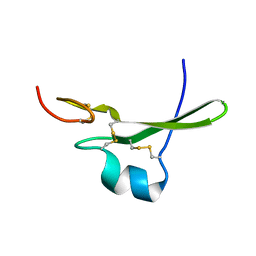 | |
3CGU
 
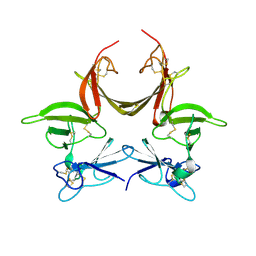 | |
7LEN
 
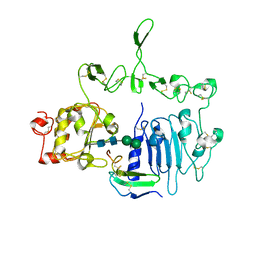 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFS
 
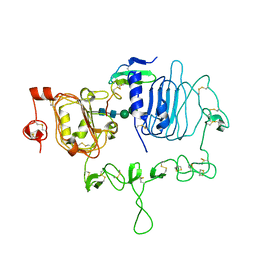 | | Crystal structure of the epidermal growth factor receptor extracellular region with A265V mutation in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 4 of Epidermal growth factor receptor, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFR
 
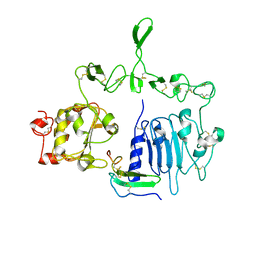 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with spermine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Proepiregulin, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7ME4
 
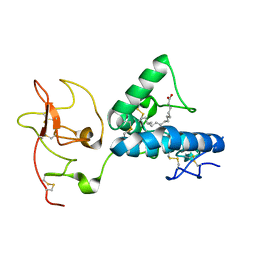 | | Structure of the extracellular WNT-binding module in Drosophila Ror2/Nrk | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOLEIC ACID, Tyrosine-protein kinase transmembrane receptor Ror2 | | Authors: | Mendrola, J.M, Shi, F, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
7ME5
 
 | | Structure of the extracellular WNT-binding module in Drl-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tyrosine-protein kinase transmembrane receptor DRL-2 | | Authors: | Shi, F, Mendrola, J.M, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
1PI1
 
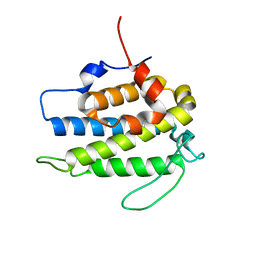 | | Crystal structure of a human Mob1 protein; toward understanding Mob-regulated cell cycle pathways. | | Descriptor: | Mob1A, ZINC ION | | Authors: | Stavridi, E.S, Harris, K.G, Huyen, Y, Bothos, J, Voewerd, P.M, Stayrook, S.E, Jeffrey, P.D, Pavletich, N.P, Luca, F.C. | | Deposit date: | 2003-05-29 | | Release date: | 2003-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a human mob1 protein. Toward understanding mob-regulated cell cycle pathways.
Structure, 11, 2003
|
|
1IHI
 
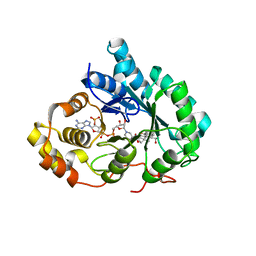 | | Crystal Structure of Human Type III 3-alpha-Hydroxysteroid Dehydrogenase/Bile Acid Binding Protein (AKR1C2) Complexed with NADP+ and Ursodeoxycholate | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, ISO-URSODEOXYCHOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jin, Y, Stayrook, S.E, Albert, R.H, Palackal, N.T, Penning, T.M, Lewis, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human type III 3alpha-hydroxysteroid dehydrogenase/bile acid binding protein complexed with NADP(+) and ursodeoxycholate.
Biochemistry, 40, 2001
|
|
1YOD
 
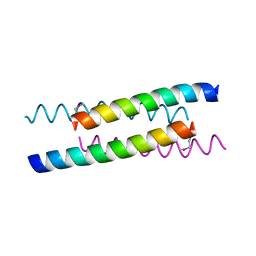 | |
1Y47
 
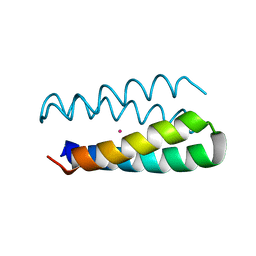 | | Structural studies of designed alpha-helical hairpins | | Descriptor: | CADMIUM ION, dueferri (DF2) | | Authors: | Lahr, S.J, Engel, D.E, Stayrook, S.E, Maglio, O, North, B, Geremia, S, Lombardi, A, DeGrado, W.F. | | Deposit date: | 2004-11-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis and design of turns in alpha-helical hairpins
J.Mol.Biol., 346, 2005
|
|
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
1M3W
 
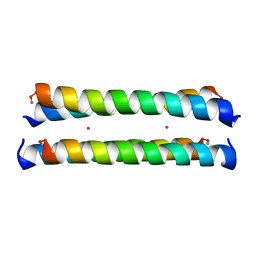 | | Crystal Structure of a Molecular Maquette Scaffold | | Descriptor: | H10H24, MERCURY (II) ION | | Authors: | Huang, S.S, Gibney, B.R, Stayrook, S.E, Dutton, P.L, Lewis, M. | | Deposit date: | 2002-07-01 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Structure of a Maquette Scaffold
J.Mol.Biol., 326, 2003
|
|
