9BAQ
 
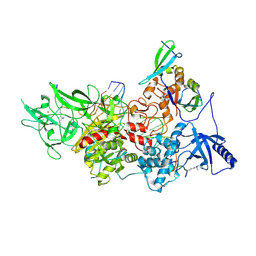 | | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3'), DNA (cytosine-5-)-methyltransferase, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex
To Be Published
|
|
9BAP
 
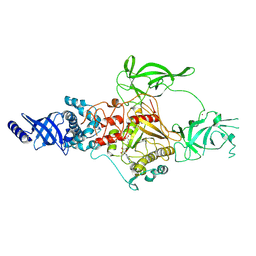 | | CryoEM structure of Apo-DIM2 | | Descriptor: | DNA (cytosine-5-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | CryoEM structure of Apo-DIM2
To Be Published
|
|
9BAZ
 
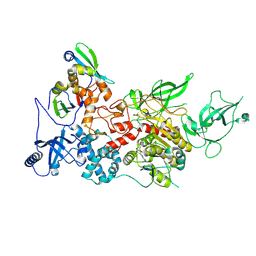 | | CryoEM structure of DIM2-HP1 complex | | Descriptor: | DNA (cytosine-5-)-methyltransferase, Heterochromatin protein one, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | CryoEM structure of DIM2-HP1 complex
To Be Published
|
|
8FYH
 
 | | G4 RNA-mediated PRC2 dimer | | Descriptor: | G4 RNA, Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Song, J, Kasinath, V. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inactivation of PRC2 by G-quadruplex RNA.
Science, 381, 2023
|
|
4DA4
 
 | |
4ZDS
 
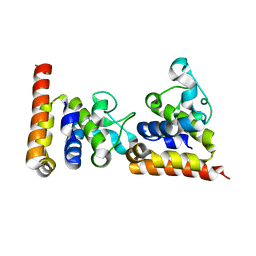 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | Descriptor: | Protein ETHYLENE INSENSITIVE 3 | | Authors: | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
1XOY
 
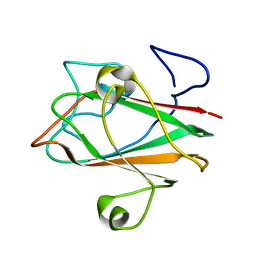 | | Solution structure of At3g04780.1, an Arabidopsis ortholog of the C-terminal domain of human thioredoxin-like protein | | Descriptor: | hypothetical protein At3g04780.1 | | Authors: | Song, J, Robert, C.T, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of At3g04780.1-des15, an Arabidopsis thaliana ortholog of the C-terminal domain of human thioredoxin-like protein.
Protein Sci., 14, 2005
|
|
1Y4O
 
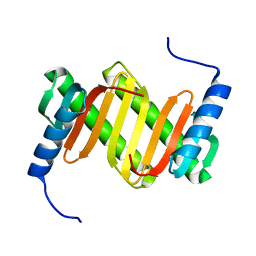 | | Solution structure of a mouse cytoplasmic Roadblock/LC7 dynein light chain | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Song, J, Tyler, R.C, Lee, M.S, Tyler, E.M, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-01 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of isoform 1 of Roadblock/LC7, a light chain in the dynein complex.
J.Mol.Biol., 354, 2005
|
|
1M8B
 
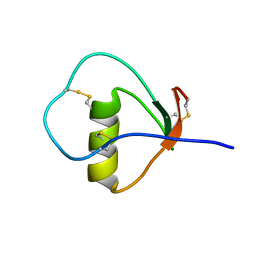 | | Solution structure of the C State of turkey ovomucoid at pH 2.5 | | Descriptor: | Ovomucoid | | Authors: | Song, J, Laskowski Jr, M, Qasim, M.A, Markley, J.L. | | Deposit date: | 2002-07-24 | | Release date: | 2002-09-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.
Biochemistry, 42, 2003
|
|
1M8C
 
 | | SOLUTION STRUCTURE OF THE T State OF TURKEY OVOMUCOID AT PH 2.5 | | Descriptor: | Ovomucoid | | Authors: | Song, J, Laskowski Jr, M, Qasim, M.A, Markley, J.L. | | Deposit date: | 2002-07-24 | | Release date: | 2002-09-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.
Biochemistry, 42, 2003
|
|
1T0G
 
 | | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold | | Descriptor: | cytochrome b5 domain-containing protein | | Authors: | Song, J, Vinarov, D.A, Tyler, E.M, Shahan, M.N, Tyler, R.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold
J.Biomol.NMR, 30, 2004
|
|
1TIZ
 
 | | Solution Structure of a Calmodulin-Like Calcium-Binding Domain from Arabidopsis thaliana | | Descriptor: | calmodulin-related protein, putative | | Authors: | Song, J, Zhao, Q, Thao, S, Frederick, R.O, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Letter to the Editor: Solution structure of a calmodulin-like calcium-binding domain from Arabidopsis thaliana
J.Biomol.NMR, 30, 2004
|
|
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
2G2B
 
 | | NMR structure of the human allograft inflammatory factor 1 | | Descriptor: | Allograft inflammatory factor 1 | | Authors: | Song, J, Tyler, R.C, Newman, C.L, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-15 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human allograft inflammatory factor 1
To be published
|
|
3WVZ
 
 | | Crystal structure of Hikeshi, a new nuclear transport receptor of Hsp70 | | Descriptor: | Protein Hikeshi | | Authors: | Song, J, Kose, S, Watanabe, A, Son, S.Y, Choi, S, Hong, R.H, Yamashita, E, Park, I.Y, Imamoto, N, Lee, S.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70s
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2JU4
 
 | | NMR structure of the gamma subunit of cGMP phosphodiesterase | | Descriptor: | (3'R)-1'-oxyl-2',2',5',5'-tetramethyl-1,3'-bipyrrolidine-2,5-dione, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma | | Authors: | Song, J, Guo, L.W, Ruoho, A.E, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-08-14 | | Release date: | 2007-10-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JWN
 
 | |
2FFT
 
 | | NMR structure of Spinach Thylakoid Soluble Phosphoprotein of 9 kDa in SDS Micelles | | Descriptor: | thylakoid soluble phosphoprotein | | Authors: | Song, J, Carlberg, I, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-12-20 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Micelle-induced folding of spinach thylakoid soluble phosphoprotein of 9 kDa and its functional implications.
Biochemistry, 45, 2006
|
|
2ETT
 
 | | Solution Structure of Human Sorting Nexin 22 PX Domain | | Descriptor: | Sorting nexin-22 | | Authors: | Song, J, Zhao, Q, Tyler, R.C, Lee, M.S, Newman, C.L, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human sorting nexin 22.
Protein Sci., 16, 2007
|
|
3WW0
 
 | | Crystal structure of F97A mutant, a new nuclear transport receptor of Hsp70 | | Descriptor: | Protein Hikeshi | | Authors: | Song, J, Kose, S, Watanabe, A, Son, S.Y, Choi, S, Hong, R.H, Yamashita, E, Park, I.Y, Imamoto, N, Lee, S.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70s
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2GW6
 
 | |
2JMU
 
 | |
3PT6
 
 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PTA
 
 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
