1LD3
 
 | | Crystal Structure of B. subilis ferrochelatase with Zn(2+) bound at the active site. | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Lecerof, D, Fodje, M.N, Leon, R.A, Olsson, U, Hansson, A, Sigfridsson, E, Ryde, U, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites
J.Biol.Inorg.Chem., 8, 2003
|
|
1N0I
 
 | | Crystal Structure of Ferrochelatase with Cadmium bound at active site | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferrochelatase, ... | | Authors: | Lecerof, D, Fodje, M.N, Leon, R.A, Olsson, U, Hansson, A, Sigfridsson, E, Ryde, U, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-10-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites
J.Biol.Inorg.Chem., 8, 2003
|
|
2XG3
 
 | | Human galectin-3 in complex with a benzamido-N-acetyllactoseamine inhibitor | | Descriptor: | BENZAMIDE, CHLORIDE ION, Galectin-3, ... | | Authors: | Diehl, C, Engstrom, O, Delaine, T, Hakansson, M, Genheden, S, Modig, K, Leffler, H, Ryde, U, Nilsson, U, Akke, M. | | Deposit date: | 2010-05-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3.
J. Am. Chem. Soc., 132, 2010
|
|
2FQL
 
 | |
6RZI
 
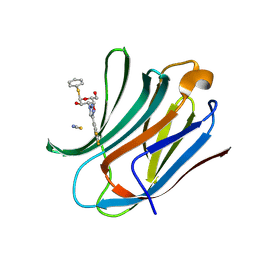 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:1(Hydrogen) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-phenylsulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
6RZJ
 
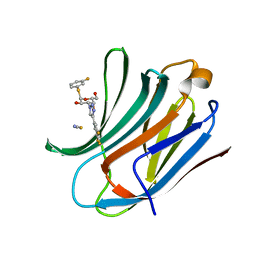 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:2(Fluorine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3-fluorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
6RZM
 
 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:5(Iodine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(3-iodanylphenyl)sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
3ZSJ
 
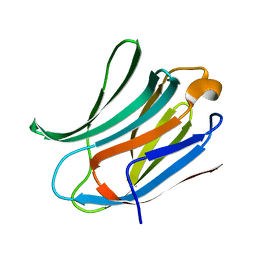 | | Crystal structure of Human Galectin-3 CRD in complex with Lactose at 0.86 angstrom resolution | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZSK
 
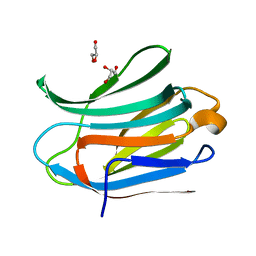 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | Descriptor: | GALECTIN-3, GLYCEROL | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZSM
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.25 angstrom resolution, at room temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZSL
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.08 angstrom resolution, at cryogenic temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
6RZH
 
 | | Galectin-3C in complex with para-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.947 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
6RZG
 
 | | Galectin-3C in complex with meta-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.015 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
6RZF
 
 | | Galectin-3C in complex with ortho-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.016 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
4EC2
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
6I76
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-3 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azido-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6I77
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-4 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azanyl-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6I78
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 5 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,5,6-tetrakis(fluoranyl)-4-(methylamino)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6I75
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,5,6-tetrakis(fluoranyl)-4-oxidanyl-phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6I74
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 1 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
7ABX
 
 | | Perdeuterated E65Q-TIM complexed with 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
7AZ9
 
 | | Perdeuterated E65Q-TIM complexed with PHOSPHOGLYCOLOHYDROXAMATE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-11-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
7AZ3
 
 | | Perdeuterated E65Q-TIM complexed with 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-11-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.15 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
7AZA
 
 | | Perdeuterated E65Q-TIM complexed with PHOSPHOGLYCOLOHYDROXAMATE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-11-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
7AZ4
 
 | | Perdeuterated E65Q-TIM complexed with 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-11-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.15 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
