2F86
 
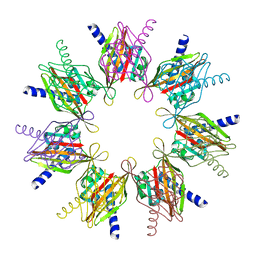 | |
3R1F
 
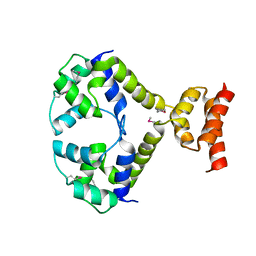 | | Crystal structure of a key regulator of virulence in Mycobacterium tuberculosis | | Descriptor: | ESX-1 secretion-associated regulator EspR | | Authors: | Rosenberg, O.S, Dovey, C, Finer-Moore, J, Stroud, R.M, Cox, J.S. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2BDW
 
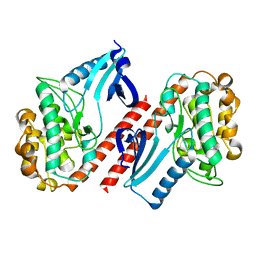 | |
4NH0
 
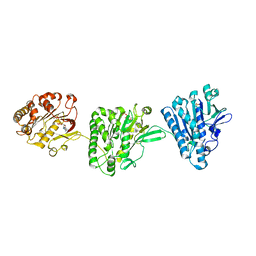 | | Cytoplasmic domain of the Thermomonospora curvata Type VII Secretion ATPase EccC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell divisionFtsK/SpoIIIE, MAGNESIUM ION, ... | | Authors: | Rosenberg, O.S, Cox, J.S, Stroud, R.M, Strauli, N, Dovala, D. | | Deposit date: | 2013-11-03 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
4M8B
 
 | | Fungal Protein | | Descriptor: | 1,2-ETHANEDIOL, Chain A of dsDNA containing the cis-regulatory element, Chain B of dsDNA containing the cis-regulatory element, ... | | Authors: | Rosenberg, O.S, Lohse, M.L, Stroud, R.M, Johnson, A.D. | | Deposit date: | 2013-08-13 | | Release date: | 2014-06-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of a new DNA-binding domain which regulates pathogenesis in a wide variety of fungi.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LYA
 
 | | EssC (ATPases 2 and 3) from Geobacillus thermodenitrificans (SeMet) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Dovala, D.L, Bendebury, A, Cox, J.S, Stroud, R.M, Rosenberg, O.S. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-04 | | Last modified: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
4LWS
 
 | | EsxA : EsxB (SeMet) hetero-dimer from Thermomonospora curvata | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Dovala, D.L, Cox, J.S, Stroud, R.M, Rosenberg, O.S. | | Deposit date: | 2013-07-28 | | Release date: | 2015-02-04 | | Last modified: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
4N1A
 
 | | Thermomonospora curvata EccC (ATPases 2 and 3) in complex with a signal sequence peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell divisionFtsK/SpoIIIE, MAGNESIUM ION, ... | | Authors: | Dovala, D.L, Bendebury, A, Cox, J.S, Stroud, R.M, Rosenberg, O.S. | | Deposit date: | 2013-10-03 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
6UMM
 
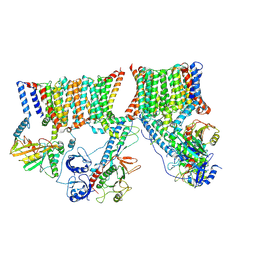 | | A complete structure of the ESX-3 translocon complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Poweleit, N, Rosenberg, O.S. | | Deposit date: | 2019-10-09 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A large inner membrane pore defines the ESX translocon
To Be Published
|
|
8CUY
 
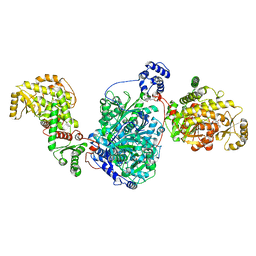 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV0
 
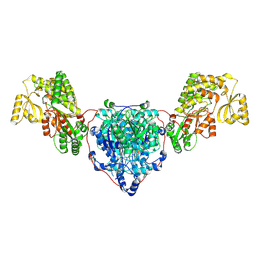 | | KS-AT domains of mycobacterial Pks13 with outward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV1
 
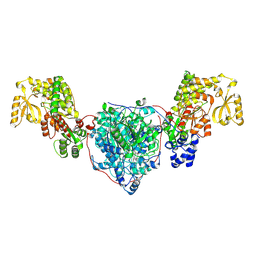 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CUZ
 
 | | KS-AT domains of mycobacterial Pks13 with inward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7UK4
 
 | | KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4MSX
 
 | | Crystal structure of an essential yeast splicing factor | | Descriptor: | ACETATE ION, Pre-mRNA-splicing factor SAD1, ZINC ION | | Authors: | Hadjivassiliou, H, Guthrie, C, Rosenberg, O.S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The crystal structure of S. cerevisiae Sad1, a catalytically inactive deubiquitinase that is broadly required for pre-mRNA splicing.
Rna, 20, 2014
|
|
5TP1
 
 | |
7R79
 
 | | Histoplasma capsulatum H88 Calcium Binding Protein 1 (Cbp1) | | Descriptor: | Calcium-binding protein | | Authors: | Herrera, N, Azimova, D, Sil, A, Rosenberg, O.S. | | Deposit date: | 2021-06-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Plos Pathog., 18, 2022
|
|
7R6U
 
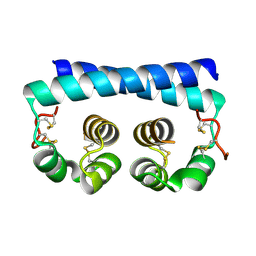 | | Paracoccidioides americana Pb03 Calcium Binding Protein 1 (Cbp1) | | Descriptor: | CBP domain-containing protein | | Authors: | Herrera, N, Azimova, D, Sil, A, Rosenberg, O.S. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Plos Pathog., 18, 2022
|
|
7KDT
 
 | |
3SOA
 
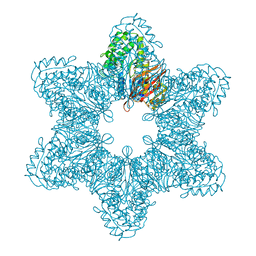 | | Full-length human CaMKII | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Calcium/calmodulin-dependent protein kinase type II subunit alpha with a beta 7 linker | | Authors: | Chao, L.H, Kuriyan, J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5501 Å) | | Cite: | A Mechanism for Tunable Autoinhibition in the Structure of a Human Ca(2+)/Calmodulin- Dependent Kinase II Holoenzyme.
Cell(Cambridge,Mass.), 146, 2011
|
|
8DV1
 
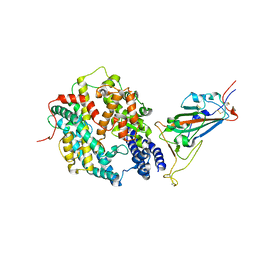 | | SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion,Immunoglobulin gamma-1 heavy chain, Spike glycoprotein | | Authors: | QCRG Structural Biology Consortium, Remesh, S.G, Merz, G.E, Brilot, A.F, Chio, U, Verba, K.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps.
Structure, 31, 2023
|
|
8DV2
 
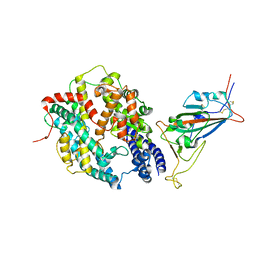 | | SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion, Spike glycoprotein | | Authors: | QCRG Structural Biology Consortium, Remesh, S.G, Merz, G.E, Brilot, A.F, Chio, U, Verba, K.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps.
Structure, 31, 2023
|
|
