1GT8
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTH
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
5AHK
 
 | | Crystal structure of acetohydroxy acid synthase Pf5 from Pseudomonas protegens | | Descriptor: | ACETOLACTATE SYNTHASE II, LARGE SUBUNIT, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Loschonsky, S, Mueller, M, Schneider, G. | | Deposit date: | 2015-02-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of the Acetohydroxy Acid Synthase Pf5 from Pseudomonas Protegens
To be Published
|
|
1H7X
 
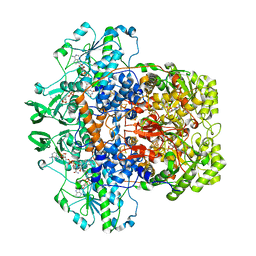 | | Dihydropyrimidine dehydrogenase (DPD) from pig, ternary complex of a mutant enzyme (C671A), NADPH and 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Dihydropyrimidine Dehydrogenase, a Major Determinant of the Pharmacokinetics of the Anti-Cancer Drug 5-Fluorouracil
Embo J., 20, 2001
|
|
2Y5T
 
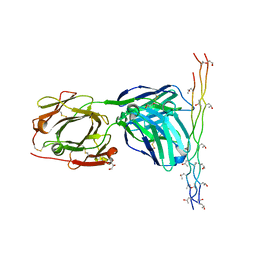 | | Crystal structure of the pathogenic autoantibody CIIC1 in complex with the triple-helical C1 peptide | | Descriptor: | C1, CHLORIDE ION, CIIC1 FAB FRAGMENT HEAVY CHAIN, ... | | Authors: | Dobritzsch, D, Lindh, I, Schneider, N, Uysal, H, Nandakumar, K.S, Burkhardt, H, Schneider, G, Holmdahl, R. | | Deposit date: | 2011-01-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Arthritogenic Anticollagen Immune Complex.
Arthritis Rheum., 63, 2011
|
|
1H7W
 
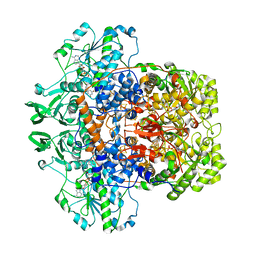 | | Dihydropyrimidine dehydrogenase (DPD) from pig | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
EMBO J., 20, 2001
|
|
4C60
 
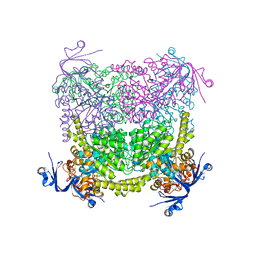 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4C5Y
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE, ZINC ION | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4C5Z
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4C65
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
5OD3
 
 | | Crystal structure of R. ruber ADH-A, mutant Y54G, L119Y | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereo- and Regioselectivity in Catalyzed Transformation of a 1,2-Disubstituted Vicinal Diol and the Corresponding Diketone by Wild Type and Laboratory Evolved Alcohol Dehydrogenases
Acs Catalysis, 8, 2018
|
|
7QXF
 
 | |
5OCX
 
 | | Crystal structure of ACPA E4 in complex with CII-C-13-CIT | | Descriptor: | 1,2-ETHANEDIOL, CII-C-13-CIT, Fab fragment anti-citrullinated protein antibody E4 - light chain, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
6FFZ
 
 | | Crystal structure of R. ruber ADH-A, mutant F43H, Y54L | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Directed Evolution of Alcohol Dehydrogenase for Improved Stereoselective Redox Transformations of 1-Phenylethane-1,2-diol and Its Corresponding Acyloin.
Biochemistry, 57, 2018
|
|
6FFX
 
 | | Crystal structure of R. ruber ADH-A, mutant F43H | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed Evolution of Alcohol Dehydrogenase for Improved Stereoselective Redox Transformations of 1-Phenylethane-1,2-diol and Its Corresponding Acyloin.
Biochemistry, 57, 2018
|
|
6FG0
 
 | | Crystal structure of R. ruber ADH-A, mutant Y54G, F43T, L119Y, F282W | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Stereo- and Regioselectivity in Catalyzed Transformation of a 1,2-Disubstituted Vicinal Diol and the Corresponding Diketone by Wild Type and Laboratory Evolved Alcohol Dehydrogenases
Acs Catalysis, 8, 2018
|
|
6FTQ
 
 | |
1ZPD
 
 | | PYRUVATE DECARBOXYLASE FROM ZYMOMONAS MOBILIS | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MONO-{4-[(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-AMINO]-2-HYDROXY-3-MERCAPTO-PENT-3-ENYL-PHOSPHONO} ESTER, ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1998-04-17 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases.
J.Biol.Chem., 273, 1998
|
|
2FTY
 
 | |
2FVK
 
 | |
2FVM
 
 | |
6SF6
 
 | |
5O8H
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A, F43H, H39Y | | Descriptor: | Alcohol dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Reddy Enugala, T, Widersten, M. | | Deposit date: | 2017-06-13 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
5O8Q
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
