3D5K
 
 | |
3RFZ
 
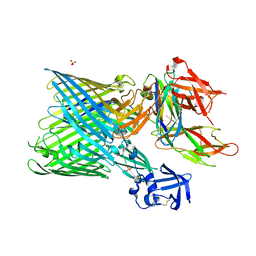 | | Crystal structure of the FimD usher bound to its cognate FimC:FimH substrate | | Descriptor: | Chaperone protein fimC, Outer membrane usher protein, type 1 fimbrial synthesis, ... | | Authors: | Phan, G, Remaut, H, Lebedev, A, Geibel, S, Waksman, G. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
5NSW
 
 | | Xenon for tunnelling analysis of the efflux pump component OprN. | | Descriptor: | Multidrug efflux outer membrane protein OprN, NICKEL (II) ION, PALMITIC ACID, ... | | Authors: | Phan, G, Prange, T, Enguene Ntsogo, Y.V, Garnier, C, Ducruix, A, Broutin, I. | | Deposit date: | 2017-04-27 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenon for tunnelling analysis of the efflux pump component OprN.
PLoS ONE, 12, 2017
|
|
2XG5
 
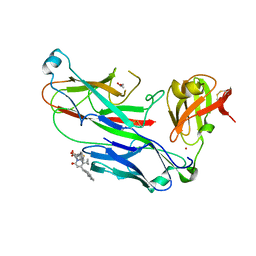 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 5d | | Descriptor: | (2R)-2-[5-CYCLOPROPYL-6-(HYDROXYSULFANYL)-4-(NAPHTHALEN-1-YLMETHYL)-2-OXOPYRIDIN-1(2H)-YL]-3-PHENYLPROPANOIC ACID, (2R,3R)-8-CYCLOPROPYL-7-(NAPHTHALEN-1-YLMETHYL)-5-OXO-2-PHENYL-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
2XG4
 
 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 2c | | Descriptor: | (3R)-8-CYCLOPROPYL-6-(MORPHOLIN-4-YLMETHYL)-7-(1-NAPHTHYLMETHYL)-5-OXO-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, COBALT (II) ION, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
6T7S
 
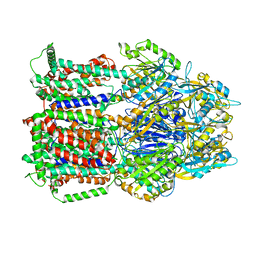 | | MexB structure solved by cryo-EM in nanodisc in absence of its protein partners | | Descriptor: | Efflux pump membrane transporter | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-23 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
6TA6
 
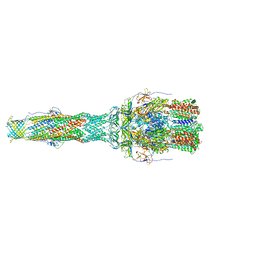 | | MexAB assembly of the Pseudomonas MexAB-OprM efflux pump reconstituted in nanodiscs | | Descriptor: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
4Y1K
 
 | |
5IUY
 
 | | Structural insights of the outer-membrane channel OprN | | Descriptor: | CHLORIDE ION, FORMIC ACID, Multidrug efflux outer membrane protein OprN, ... | | Authors: | Ntsogo, Y, Garnier, C, Phan, G, Monlezun, L, Benas, P, Broutin, I. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Xenon for tunnelling analysis of the efflux pump component OprN.
PLoS ONE, 12, 2017
|
|
6TA5
 
 | | OprM-MexA complex from the MexAB-OprM Pseudomonas aeruginosa whole assembly reconstituted in nanodiscs | | Descriptor: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
6ZRE
 
 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, SULFATE ION, ... | | Authors: | Ntsogo Enguene, Y.V, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Deciphering the role of OprM constrictions in the opening mechanism of the MexAB-OprM efflux pump from Pseudomonas aeruginosa.
To Be Published
|
|
7AKZ
 
 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, octyl beta-D-glucopyranoside | | Authors: | Ntsogo Enguene, V.Y, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-10-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa
To Be Published
|
|
8Q8O
 
 | |
2C1N
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
2C1J
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
3OHN
 
 | | Crystal structure of the FimD translocation domain | | Descriptor: | Outer membrane usher protein FimD | | Authors: | Wang, T, Li, H. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
7OLI
 
 | | Crystal structure of Pab-AGOG in complex with 8-oxoguanosine | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-8-OXOGUANOSINE, N-glycosylase/DNA lyase | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OLB
 
 | | Crystal structure of Pab-AGOG, an 8-oxoguanine DNA glycosylase from Pyrococcus abyssi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OUE
 
 | | Crystal structure of a trapped Pab-AGOG/single-standed DNA covalent intermediate | | Descriptor: | DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), N-glycosylase/DNA lyase, PHOSPHATE ION, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OU3
 
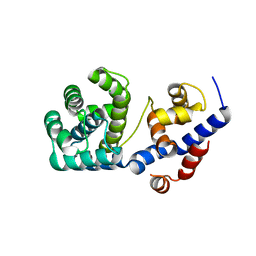 | | Crystal structure of Tga-AGOG, an 8-oxoguanine DNA glycosylase from Thermococcus gammatolerans | | Descriptor: | CHLORIDE ION, GLYCEROL, N-glycosylase/DNA lyase | | Authors: | Coste, F, Confalonieri, F, Castaing, B. | | Deposit date: | 2021-06-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P0W
 
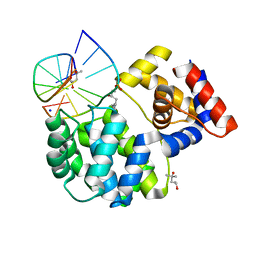 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing thymine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OY7
 
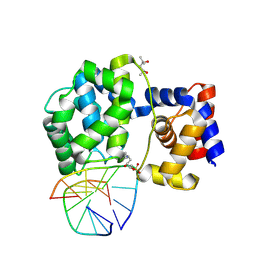 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing cytosine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P8L
 
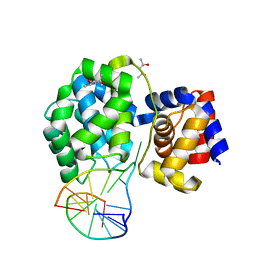 | | Crystal structure of Pyrococcus abyssi 8-oxoguanine DNA glycosylase (PabAGOG) in complex with dsDNA containing cytosine opposite to 8-oxoG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(8OG)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Flament, D, Castaing, B. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P9Z
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing adenine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
3ME3
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-{[4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-1,4-diazepan-1-yl]sulfonyl}aniline, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
