3L4C
 
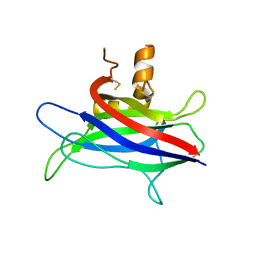 | | Structural basis of membrane-targeting by Dock180 | | Descriptor: | BETA-MERCAPTOETHANOL, Dedicator of cytokinesis protein 1 | | Authors: | Premkumar, L, Bobkov, A.A, Patel, M, Jaroszewski, L, Bankston, L.A, Stec, B, Vuori, K, Cote, J.-F, Liddington, R.C. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs).
J.Biol.Chem., 285, 2010
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
7EWO
 
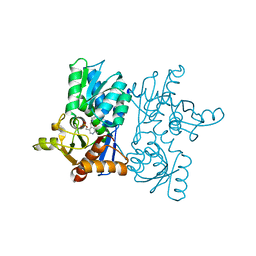 | | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase | | Authors: | Rahisuddin, R, Ekka, M.K, Singh, A.K, Saini, N, Patel, M, Kumar, N, Kumaran, S. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae
To Be Published
|
|
1TXT
 
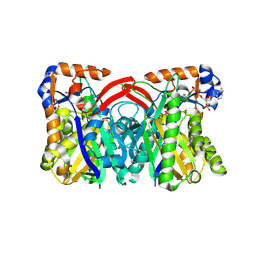 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1TVZ
 
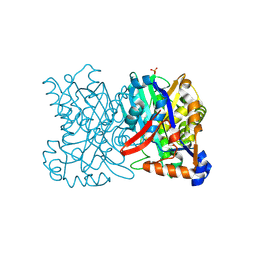 | | Crystal structure of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Staphylococcus aureus | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, SULFATE ION | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1D9E
 
 | | STRUCTURE OF E. COLI KDO8P SYNTHASE | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Radaev, S, Dastidar, P, Patel, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 1999-10-27 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of 3-deoxy-D-manno-octulosonate 8-phosphate synthase.
J.Biol.Chem., 275, 2000
|
|
2VSZ
 
 | | Crystal Structure of the ELMO1 PH domain | | Descriptor: | ENGULFMENT AND CELL MOTILITY PROTEIN 1 | | Authors: | Komander, D, Patel, M, Barford, D, Cote, J.-F. | | Deposit date: | 2008-05-01 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Alpha-Helical Extension of the Elmo1 Pleckstrin Homology Domain Mediates Direct Interaction to Dock180 and is Critical in Rac Signaling.
Molecular Biology of the Cell, 19, 2008
|
|
2N9C
 
 | | NRAS Isoform 5 | | Descriptor: | GTPase NRas | | Authors: | Markowitz, J, Mal, T.K, Yuan, C, Courtney, N.B, Patel, M, Stiff, A.R, Blachly, J, Walker, C, Eisfeld, A, de la Chapelle, A, Carson III, W.E. | | Deposit date: | 2015-11-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of NRAS isoform 5.
Protein Sci., 25, 2016
|
|
5TWD
 
 | | CTX-M-14 P167S apoenzyme | | Descriptor: | Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5U53
 
 | | CTX-M-14 E166A with acylated ceftazidime molecule | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase, NITRATE ION | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-12-06 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TWE
 
 | | CTX-M-14 P167S:S70G mutant enzyme crystallized with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TW6
 
 | | CTX-M-14 P167S:E166A mutant with acylated ceftazidime molecule | | Descriptor: | 1,2-ETHANEDIOL, ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-11 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
6DBP
 
 | |
5VTH
 
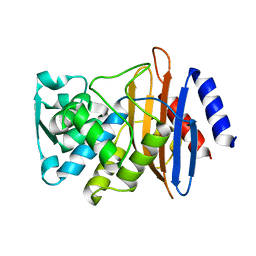 | | CTX-M-14 P167S:E166A mutant | | Descriptor: | Beta-lactamase | | Authors: | Hu, L, Patel, M, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2017-05-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
6DE7
 
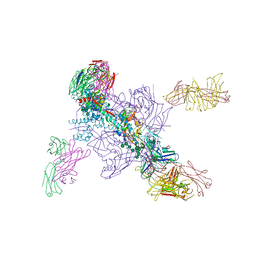 | |
7DJQ
 
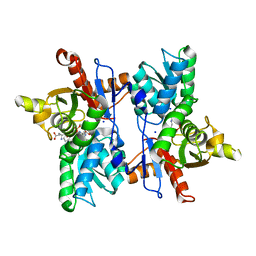 | |
6X9I
 
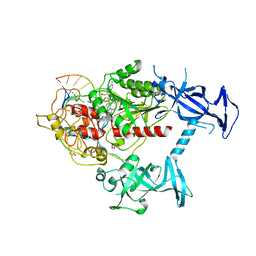 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X9K
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3685032A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]sulfanyl}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X9J
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830052 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
8FIU
 
 | | Potent long-acting inhibitors targeting HIV-1 capsid based on a versatile quinazolin-4-one scaffold | | Descriptor: | 1,2-ETHANEDIOL, HIV-1 capsid, N-[(1S)-1-{(3P)-3-{4-chloro-3-[(methanesulfonyl)amino]-1-methyl-1H-indazol-7-yl}-7-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-4-oxo-3,4-dihydroquinazolin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Nolte, R.T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent Long-Acting Inhibitors Targeting the HIV-1 Capsid Based on a Versatile Quinazolin-4-one Scaffold.
J.Med.Chem., 66, 2023
|
|
3IOX
 
 | | Crystal Structure of A3VP1 of AgI/II of Streptococcus mutans | | Descriptor: | AgI/II, CALCIUM ION, SULFITE ION, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Patel, M, Robinette, R, Crowley, P, Michalek, S.M, Brady, L.J, Deivanayagam, C.C. | | Deposit date: | 2009-08-14 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IPK
 
 | | Crystal Structure of A3VP1 of AgI/II of Streptococcus mutans | | Descriptor: | AgI/II, CALCIUM ION, SULFATE ION, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Patel, M, Robinette, R, Crowley, P, Michalek, S.M, Brady, L.J, Deivanayagam, C.C. | | Deposit date: | 2009-08-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ZD2
 
 | | THE STRUCTURE OF THE TWO N-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 1 SHOWS FORMATION OF A NOVEL DIMERISATION INTERFACE | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 1 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZD1
 
 | | STRUCTURE OF THE TWO C-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 2 | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 2 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6YUW
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | Descriptor: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
