1OIX
 
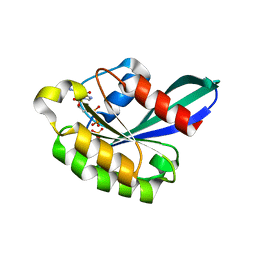 | | X-ray structure of the small G protein Rab11a in complex with GDP and Pi | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Evidence for Substrate-Assisted GTP Hydrolysis by a Small GTP Binding Protein
Structure, 13, 2005
|
|
2J5X
 
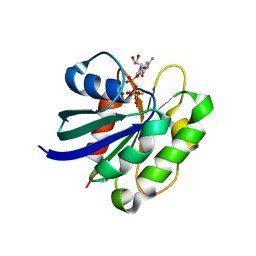 | | STRUCTURE OF THE SMALL G PROTEIN ARF6 IN COMPLEX WITH GTPGAMMAS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-RIBOSYLATION FACTOR 6, MAGNESIUM ION | | Authors: | Pasqualato, S, Menetrey, J, Franco, M, Cherfils, J. | | Deposit date: | 2006-09-20 | | Release date: | 2006-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structural Gdp-GTP Cycle of Human Arf6.
Embo Rep., 2, 2001
|
|
1OIW
 
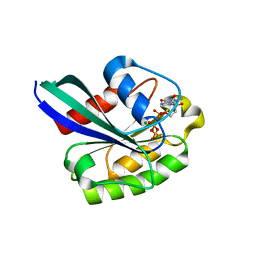 | | X-ray structure of the small G protein Rab11a in complex with GTPgammaS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, RAS-RELATED PROTEIN RAB-11A | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structural Gdp/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes
J.Biol.Chem., 279, 2004
|
|
1OIV
 
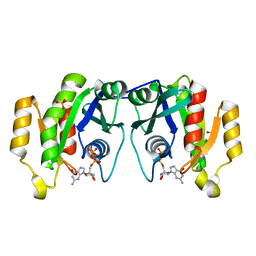 | | X-ray structure of the small G protein Rab11a in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, RAS-RELATED PROTEIN RAB-11A, ... | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structural Gdp/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes
J.Biol.Chem., 279, 2004
|
|
6TE1
 
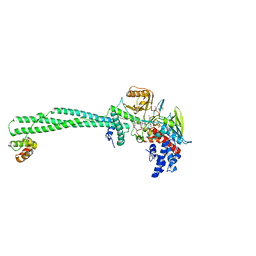 | | Structure of the KDM1A/CoREST complex with the inhibitor 2-[3-{4-chloro-3-[(4-chlorophenyl)ethynyl]phenyl}-1-(3-morpholin-4-ylpropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]-2-oxoethanol | | Descriptor: | 5-[4-cyclobutyl-1-[2-(4-piperidin-4-yloxyphenoxy)ethyl]imidazol-2-yl]-4-methyl-thieno[3,2-b]pyrrole, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Pasqualato, S, Cecatiello, V. | | Deposit date: | 2019-11-11 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of Reversible Inhibitors of KDM1A Efficacious in Acute Myeloid Leukemia Models.
Acs Med.Chem.Lett., 11, 2020
|
|
6HC2
 
 | | Crystal structure of NuMA/LGN hetero-hexamers | | Descriptor: | G-protein-signaling modulator 2, Nuclear mitotic apparatus protein 1 | | Authors: | Pasqualato, S, Culurgioni, S, Foadi, J, Alfieri, A, Mapelli, M. | | Deposit date: | 2018-08-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions.
Nat Commun, 10, 2019
|
|
1E7Z
 
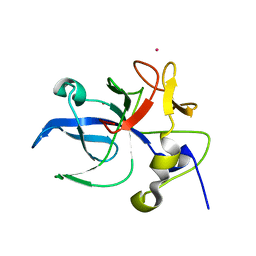 | | Crystal structure of the EMAP2/RNA binding domain of the p43 protein from human aminoacyl-tRNA synthetase complex | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II, MERCURY (II) ION | | Authors: | Pasqualato, S, Kerjan, P, Renault, L, Menetrey, J, Mirande, M, Cherfils, J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Emapii Domain of Human Aminoacyl-tRNA Synthetase Complex Reveals Evolutionary Dimeric Mimicry
Embo J., 20, 2001
|
|
2VE7
 
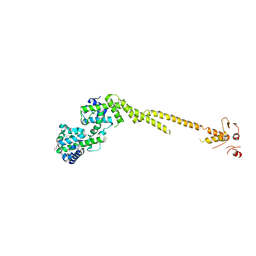 | | Crystal structure of a bonsai version of the human Ndc80 complex | | Descriptor: | GLYCEROL, KINETOCHORE PROTEIN HEC1, KINETOCHORE PROTEIN SPC25, ... | | Authors: | Ciferri, C, Pasqualato, S, Dos Reis, G, Screpanti, E, Maiolica, A, Polka, J, De Luca, J.G, De Wulf, P, Salek, M, Rappsilber, J, Moores, C.A, Salmon, E.D, Musacchio, A. | | Deposit date: | 2007-10-17 | | Release date: | 2008-05-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Implications for Kinetochore-Microtubule Attachment from the Structure of an Engineered Ndc80 Complex
Cell(Cambridge,Mass.), 133, 2008
|
|
4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
6QJA
 
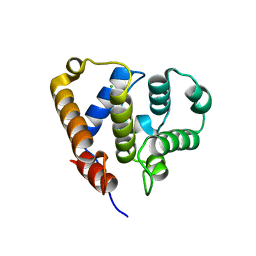 | | Organizational principles of the NuMA-Dynein interaction interface and implications for mitotic spindle functions | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nuclear mitotic apparatus protein 1 | | Authors: | Renna, C, Rizzelli, F, Carminati, M, Gaddoni, C, Pirovano, L, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2019-01-23 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Organizational Principles of the NuMA-Dynein Interaction Interface and Implications for Mitotic Spindle Functions.
Structure, 28, 2020
|
|
3IZ0
 
 | | Human Ndc80 Bonsai Decorated Microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Ramey, V.H, Pasqualato, S, Ball, D.A, Grigorieff, N, Musacchio, A, Nogales, E. | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | The Ndc80 kinetochore complex forms oligomeric arrays along microtubules.
Nature, 467, 2010
|
|
1KU1
 
 | | Crystal Structure of the Sec7 Domain of Yeast GEA2 | | Descriptor: | ARF guanine-nucleotide exchange factor 2 | | Authors: | Renault, L, Christova, P, Guibert, B, Pasqualato, S, Cherfils, J. | | Deposit date: | 2002-01-20 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of domain closure of Sec7 domains and role in BFA sensitivity.
Biochemistry, 41, 2002
|
|
4BE8
 
 | | NEDD4 HECT A889F structure | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE NEDD4 | | Authors: | Maspero, E, Valentini, E, Mari, S, Cecatiello, V, Polo, S, Pasqualato, S. | | Deposit date: | 2013-03-06 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | Structure of a Ubiquitin-Loaded Hect Ligase Reveals the Molecular Basis for Catalytic Priming
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BBN
 
 | | NEDD4 HECT-Ub:Ub complex | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE NEDD4, POLYUBIQUITIN-B | | Authors: | Maspero, E, Valentini, E, Mari, S, Cecatiello, V, Polo, S, Pasqualato, S. | | Deposit date: | 2012-09-27 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of a Ubiquitin-Loaded Hect Ligase Reveals the Molecular Basis for Catalytic Priming
Nat.Struct.Mol.Biol., 20, 2013
|
|
2XBF
 
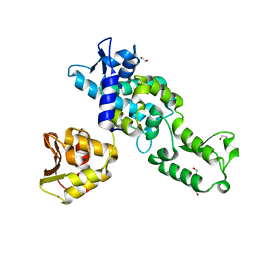 | | Nedd4 HECT structure | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE NEDD4 | | Authors: | Maspero, E, Cecatiello, V, Musacchio, A, Polo, S, Pasqualato, S. | | Deposit date: | 2010-04-09 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structure of the Hect:Ubiquitin Complex and its Role in Ubiquitin Chain Elongation
Embo Rep., 12, 2011
|
|
5LHG
 
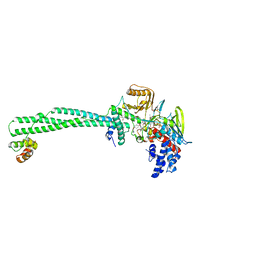 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-methyl-N-[4-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-methyl-N-[4-[[4-[(1-methyl-4-piperidyl)oxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LHI
 
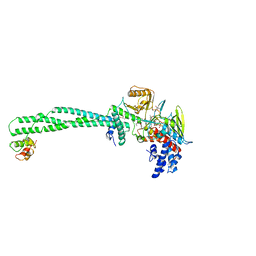 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
2XBB
 
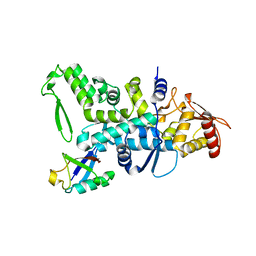 | | Nedd4 HECT:Ub complex | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE NEDD4, GLYCEROL, UBIQUITIN | | Authors: | Maspero, E, Cecatiello, V, Musacchio, A, Polo, S, Pasqualato, S. | | Deposit date: | 2010-04-08 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure of the Hect:Ubiquitin Complex and its Role in Ubiquitin Chain Elongation
Embo Rep., 12, 2011
|
|
5LHH
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-ethyl-N-[3-(methoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-ethyl-~{N}-[3-(methoxymethyl)-2-[[4-[[(3~{R})-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
1FL0
 
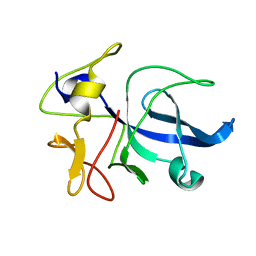 | | CRYSTAL STRUCTURE OF THE EMAP2/RNA-BINDING DOMAIN OF THE P43 PROTEIN FROM HUMAN AMINOACYL-TRNA SYNTHETASE COMPLEX | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II | | Authors: | Renault, L, Kerjan, P, Pasqualato, S, Menetrey, J, Robinson, J.-C, Kawaguchi, S, Vassylyev, D.G, Yokoyama, S, Mirande, M, Cherfils, J. | | Deposit date: | 2000-08-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry.
EMBO J., 20, 2001
|
|
5FJH
 
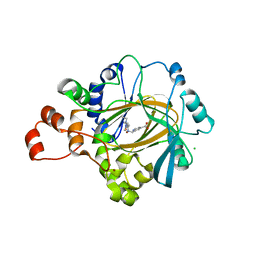 | |
5FJK
 
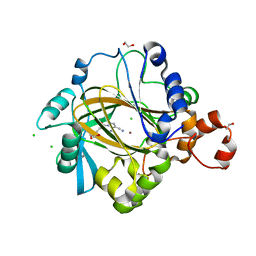 | | Crystal structure of human JMJD2C catalytic domain in complex 6-ethyl- 5-methyl-7-oxo-4,7-dihydropyrazolo(1,5-a)pyrimidine-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 6-ethyl-5-methyl-7-oxidanylidene-1H-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CHLORIDE ION, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2015-10-09 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structures of Human Jmjd2C Catalytic Domain Bound to Inhibitors
To be Published
|
|
6ZT0
 
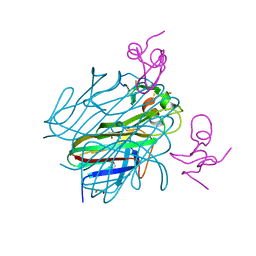 | | Crystal structure of the Eiger TNF domain/Grindelwald extracellular domain complex | | Descriptor: | 1,2-ETHANEDIOL, Protein eiger, Protein grindelwald | | Authors: | Palmerini, V, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions.
Nat Commun, 12, 2021
|
|
6ZSZ
 
 | |
6ZSY
 
 | |
