5O78
 
 | |
5O9C
 
 | |
8QXC
 
 | | Crystal structure of antibody Fab MIL-3 with PenG-Lys | | Descriptor: | (2R,4S)-2-[(1R)-2-[[(5S)-5-acetamido-6-oxidanyl-6-oxidanylidene-hexyl]amino]-2-oxidanylidene-1-(2-phenylethanoylamino)ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Heavy chain of FAB MIL-3, Light chain of FAB MIL-3 | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalent penicillin-protein conjugates elicit anti-drug antibodies that are clonally and functionally restricted.
Nat Commun, 15, 2024
|
|
7OAO
 
 | | Nanobody C5 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAY
 
 | | Nanobody F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAQ
 
 | | Nanobody H3 AND C1 bound to RBD with Kent mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAU
 
 | | Nanobody C5 bound to Kent variant RBD (N501Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5, GLYCEROL, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAP
 
 | | Nanobody H3 AND C1 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAN
 
 | | Nanobody C5 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Weckener, M. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
2BI7
 
 | | udp-galactopyranose mutase from Klebsiella pneumoniae oxidised FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Srikannathasan, V, Naismith, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Mycobacteria Tuberculosis and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the Oxidised State and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the (Active) Reduced State.
J.Mol.Biol., 348, 2005
|
|
5FNV
 
 | | a new complex structure of tubulin with an alpha-beta unsaturated lactone | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, Y, Naismith, J, Zhu, X. | | Deposit date: | 2015-11-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Pironetin Reacts Covalently with Cysteine-316 of Alpha-Tubulin to Destabilize Microtubule.
Nat.Commun., 7, 2016
|
|
2JKC
 
 | | Crystal Structure of E346D of Tryptophan 7-Halogenase (PrnA) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Zhu, X, Naismith, J.H. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the mechanism of enzymatic chlorination of tryptophan.
Angew. Chem. Int. Ed. Engl., 47, 2008
|
|
2XVO
 
 | | SSO1725, a protein involved in the CRISPR/Cas pathway | | Descriptor: | BETA-MERCAPTOETHANOL, SSO1725, SULFATE ION | | Authors: | Reeks, J, Liu, H, Naismith, J, White, M, McMahon, S. | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-29 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mechanism of the Cmr Complex for Crispr-Mediated Antiviral Immunity.
Mol.Cell, 45, 2012
|
|
4AG3
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with NADPH at 1.8A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | Authors: | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | Deposit date: | 2012-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4AFN
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa at 2.3A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, PENTAETHYLENE GLYCOL | | Authors: | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4AI4
 
 | |
4BO4
 
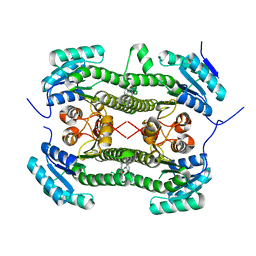 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2-methoxyphenyl)-3,4- dihydro-2H-quinoline-1-carboxamide at 2.7A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2-methoxyphenyl)-3,4-dihydro-2H-quinoline-1-carboxamide | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNX
 
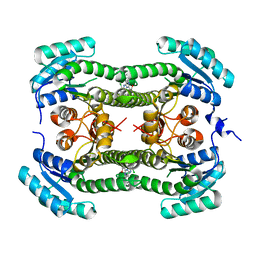 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 6-(4-(2-chloroanilino)- 1H-quinazolin-2-ylidene)cyclohexa-2, 4-dien-1-one at 2.3A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 6-[4-(2-chloroanilino)-1H-quinazolin-2-ylidene]cyclohexa-2,4-dien-1-one | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO3
 
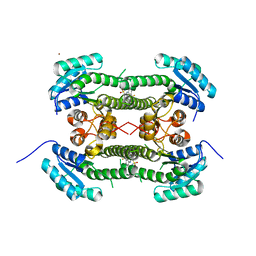 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2-(3-(trifluoromethyl) anilino)pyridine-3-sulfonamide at 2.5A resolution | | Descriptor: | 2-(3-(trifluoromethyl)anilino)pyridine-3-sulfonamide, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NICKEL (II) ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNU
 
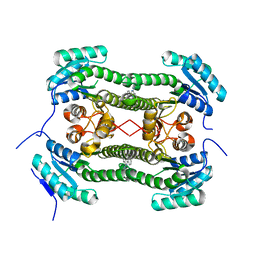 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2-phenyl-4-(1,2,4- triazol-4-yl)quinazoline at 2.0A resolution | | Descriptor: | 2-phenyl-4-(1,2,4-triazol-4-yl)quinazoline, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO1
 
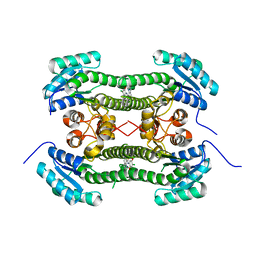 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(4-chloro-2,5- dimethoxyphenyl)quinoline-8-carboxamide at 2.2A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(4-CHLORO-2,5-DIMETHOXYPHENYL)QUINOLINE-8-CARBOXAMIDE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO9
 
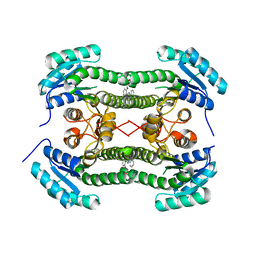 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 5-(2-(furan-2-ylmethoxy) phenyl)-2-phenyltetrazole at 2.9A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 5-[2-(FURAN-2-YLMETHOXY)PHENYL]-2-PHENYLTETRAZOLE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNW
 
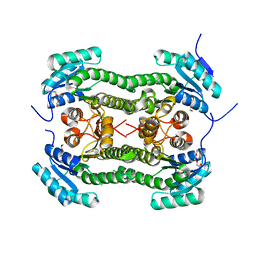 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with an unknown ligand at 1. 6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO5
 
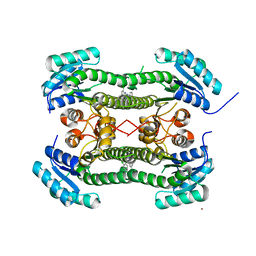 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2-chlorophenyl)-4- pyrrol-1-yl-1,3,5-triazin-2-amine at 2.6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2-chlorophenyl)-4-pyrrol-1-yl-1,3,5-triazin-2-amine, NICKEL (II) ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNV
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(2-chlorophenyl)-3-(1- methylbenzimidazol-2-yl)urea at 2.5A resolution | | Descriptor: | 1-(2-chlorophenyl)-3-(1-methylbenzimidazol-2-yl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in Resolution
Acs Chem.Biol., 8, 2013
|
|
