1JM4
 
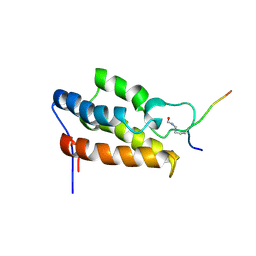 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
1JSP
 
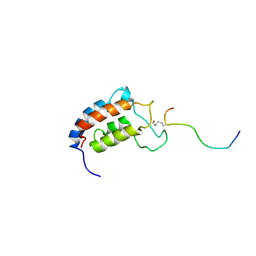 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | Descriptor: | CREB-BINDING PROTEIN, tumor protein p53 | | Authors: | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | Deposit date: | 2001-08-17 | | Release date: | 2002-08-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
2RO1
 
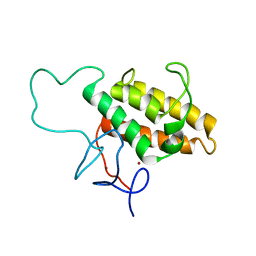 | | NMR Solution Structures of Human KAP1 PHD finger-bromodomain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Zeng, L, Yap, K.L, Ivanov, A.V, Wang, X, Mujtaba, S, Plotnikova, O, Rauscher, F.J. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing
Nat.Struct.Mol.Biol., 15, 2008
|
|
1WUG
 
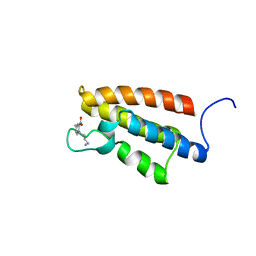 | | complex structure of PCAF bromodomain with small chemical ligand NP1 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-4-METHYL-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-07 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1WUM
 
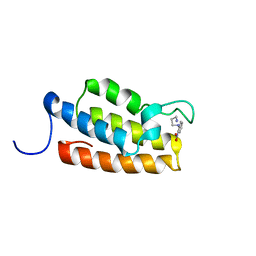 | | Complex structure of PCAF bromodomain with small chemical ligand NP2 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1XR0
 
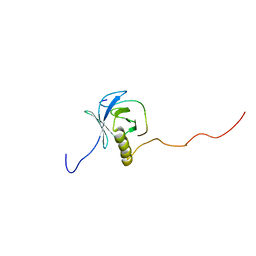 | | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors | | Descriptor: | Basic fibroblast growth factor receptor 1, FGFR signalling adaptor SNT-1 | | Authors: | Dhalluin, C, Yan, K.S, Plotnikova, O, Lee, K.W, Zeng, L, Kuti, M, Mujtaba, S, Goldfarb, M.P, Zhou, M.-M. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors
Mol.Cell, 6, 2000
|
|
2L85
 
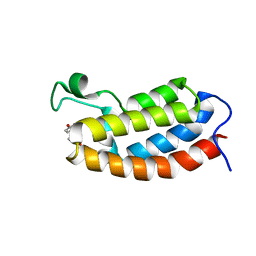 | | Solution NMR structures of CBP bromodomain with small molecule of HBS | | Descriptor: | 4-[(E)-(4-hydroxyphenyl)diazenyl]benzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
2L84
 
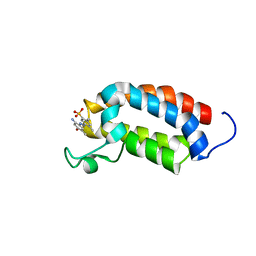 | | Solution NMR structures of CBP bromodomain with small molecule j28 | | Descriptor: | 5-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-2,4-dimethylbenzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
2D82
 
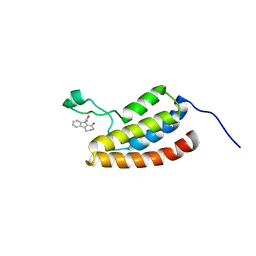 | | Target Structure-Based Discovery of Small Molecules that Block Human p53 and CREB Binding Protein (CBP) Association | | Descriptor: | 9-ACETYL-2,3,4,9-TETRAHYDRO-1H-CARBAZOL-1-ONE, CREB-binding protein | | Authors: | Sachchidanand, Resnick-Silverman, L, Yan, S, Mujtaba, S, Liu, W.J, Zeng, L, Manfredi, J.J, Zhou, M.M. | | Deposit date: | 2005-12-01 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Target structure-based discovery of small molecules that block human p53 and CREB binding protein association
Chem.Biol., 13, 2006
|
|
2KVM
 
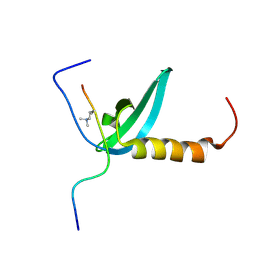 | |
4NUC
 
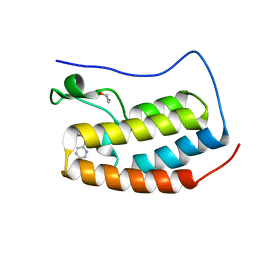 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS435 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(4-hydroxy-3,5-dimethylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
4NUE
 
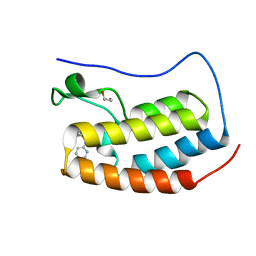 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS267 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(2-amino-4-hydroxy-3,5-dimethylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
4NUD
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS436 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
1HZM
 
 | |
2MFQ
 
 | |
