4R8P
 
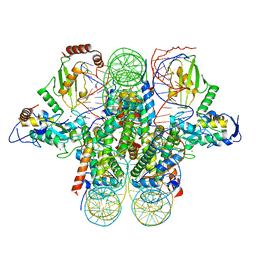 | | Crystal structure of the Ring1B/Bmi1/UbcH5c PRC1 ubiquitylation module bound to the nucleosome core particle | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase RING2, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | McGinty, R.K, Henrici, R.C, Tan, S. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2846 Å) | | Cite: | Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome.
Nature, 514, 2014
|
|
6NN6
 
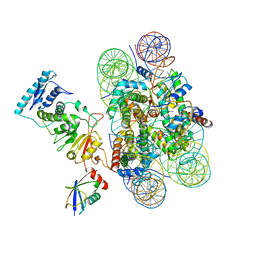 | | Structure of Dot1L-H2BK120ub nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Anderson, C.J, Baird, M.R, Hsu, A, Barbour, E.H, Koyama, Y, Borgnia, M.J, McGinty, R.K. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase.
Cell Rep, 26, 2019
|
|
7TAN
 
 | | Structure of VRK1 C-terminal tail bound to nucleosome core particle | | Descriptor: | Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, Histone H3.2, ... | | Authors: | Spangler, C.J, Budziszewski, G.R, McGinty, R.K. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity.
Nucleic Acids Res., 50, 2022
|
|
7M5U
 
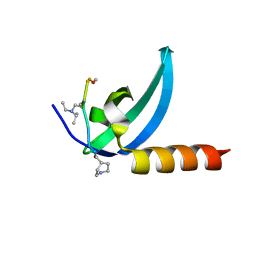 | | Crystal structure of human MPP8 chromodomain in complex with peptidomimetic ligand UNC5246 | | Descriptor: | M-phase phosphoprotein 8, UNC5246 | | Authors: | Budziszewski, G.R, McGinty, R.K, Waybright, J.M, Norris, J.L, James, L.I. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Peptidomimetic Ligand Targeting the Chromodomain of MPP8 Reveals HRP2's Association with the HUSH Complex.
Acs Chem.Biol., 16, 2021
|
|
5HQ2
 
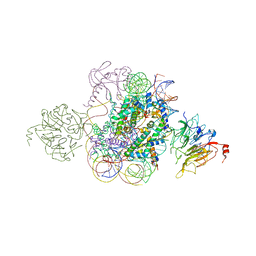 | | Structural model of Set8 histone H4 Lys20 methyltransferase bound to nucleosome core particle | | Descriptor: | DNA (149-MER), Guanine nucleotide exchange factor SRM1, Histone H2A, ... | | Authors: | Tavarekere, G, McGinty, R.K, Tan, S. | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Multivalent Interactions by the Set8 Histone Methyltransferase With Its Nucleosome Substrate.
J.Mol.Biol., 428, 2016
|
|
7UV9
 
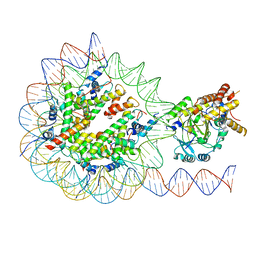 | | KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate | | Descriptor: | DNA (185-MER), FE (III) ION, Histone H2A type 1, ... | | Authors: | Spangler, C.J, Skrajna, A, Foley, C.A, Budziszewski, G.R, Azzam, D.N, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
7UVA
 
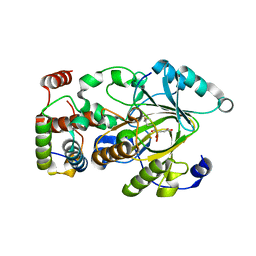 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
7JO9
 
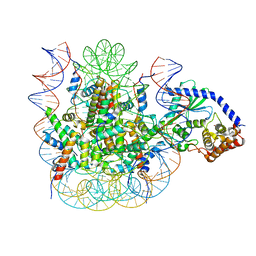 | | 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
7JOA
 
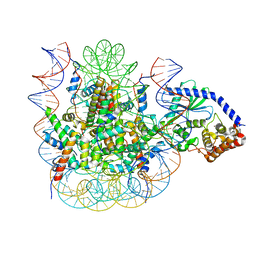 | | 2:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
3QZT
 
 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QZS
 
 | |
3QZV
 
 | | Crystal Structure of BPTF PHD-linker-bromo in complex with histone H4K12ac peptide | | Descriptor: | Histone H4, Nucleosome-remodeling factor subunit BPTF, ZINC ION | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
