3QHT
 
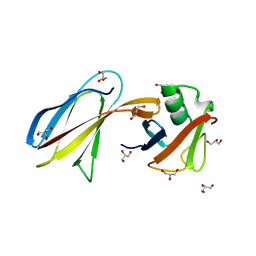 | | Crystal Structure of the Monobody ySMB-1 bound to yeast SUMO | | Descriptor: | GLYCEROL, Monobody ySMB-1, Ubiquitin-like protein SMT3 | | Authors: | Koide, S, Gilbreth, R.N. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7TVJ
 
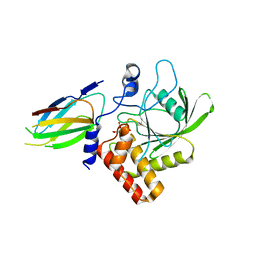 | | Crystal Structure of Monobody Mb(SHP2PTP_13)/SHP2 PTP Domain Complex | | Descriptor: | CITRATE ANION, Mb(SHP2PTP_13), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Monobody Inhibitor Selective to the Phosphatase Domain of SHP2 and its Use as a Probe for Quantifying SHP2 Allosteric Regulation.
J.Mol.Biol., 435, 2023
|
|
7LO8
 
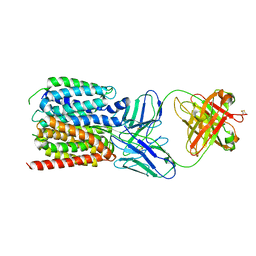 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO7
 
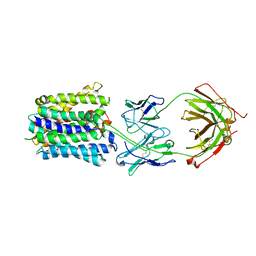 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
5KVM
 
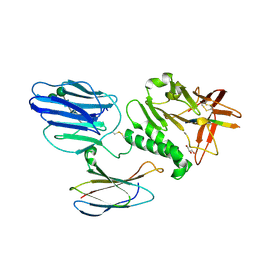 | | Extracellular region of mouse GPR56/ADGRG1 in complex with FN3 monobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor G1, ... | | Authors: | Salzman, G.S, Ding, C, Koide, S, Arac, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structural Basis for Regulation of GPR56/ADGRG1 by Its Alternatively Spliced Extracellular Domains.
Neuron, 91, 2016
|
|
8EZG
 
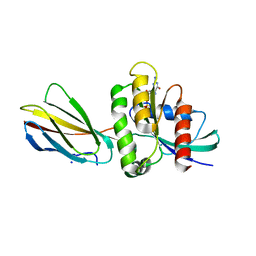 | | Monobody 12D1 bound to KRAS(G12D) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-10-31 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F0M
 
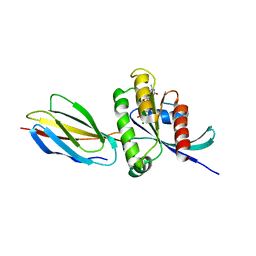 | | Monobody 12D5 bound to KRAS(G12D) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2FKJ
 
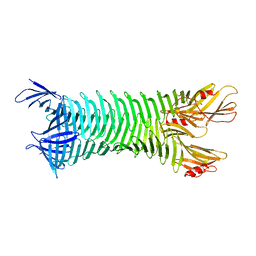 | | The crystal structure of engineered OspA | | Descriptor: | Outer Surface Protein A | | Authors: | Makabe, K, Terechko, V, Gawlak, G, Yan, S, Koide, S. | | Deposit date: | 2006-01-04 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AF5
 
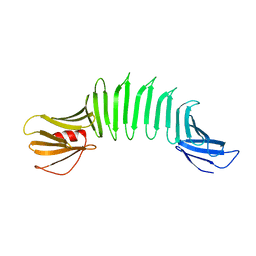 | | 2.5A X-ray Structure of Engineered OspA protein | | Descriptor: | Engineered Outer Surface Protein A (OspA) with the inserted two beta-hairpins | | Authors: | Makabe, K, Mcelheny, D, Tereshko, V, Hilyard, A, Koide, A, Koide, S. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3UYO
 
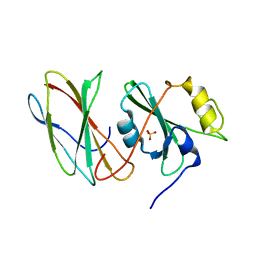 | |
2PI3
 
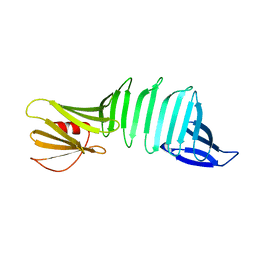 | |
3T04
 
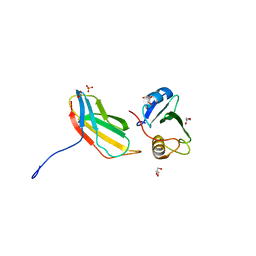 | | Crystal structure of monobody 7c12/abl1 sh2 domain complex | | Descriptor: | GLYCEROL, MONOBODY 7C12, SULFATE ION, ... | | Authors: | Wojcik, J.B, Wyrzucki, A.M, Koide, S. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the SH2-Kinase Interface in Bcr-Abl Inhibits Leukemogenesis.
Cell(Cambridge,Mass.), 147, 2011
|
|
2QBW
 
 | | The crystal structure of PDZ-Fibronectin fusion protein | | Descriptor: | PDZ-Fibronectin fusion protein, Polypeptide | | Authors: | Huang, J, Makabe, K, Koide, A, Koide, S. | | Deposit date: | 2007-06-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of protein function leaps by directed domain interface evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4M1C
 
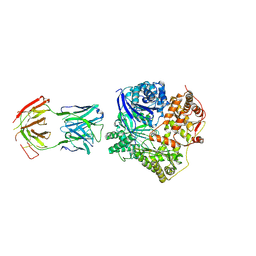 | | Crystal Structure Analysis of Fab-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Amyloid-Beta (1-40) | | Descriptor: | Amyloid beta A4 protein, Fab-bound IDE, heavy chain, ... | | Authors: | McCord, L.M, Liang, W, Farcasanu, M, Scherpelz, K, Meredith, S.C, Koide, S, Tang, W.J. | | Deposit date: | 2013-08-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5007 Å) | | Cite: | Crystal Structure Analysis of Fab-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Amyloid-Beta (1-40)
To be Published
|
|
5VG9
 
 | | Structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase) without a monobody | | Descriptor: | Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
5V7P
 
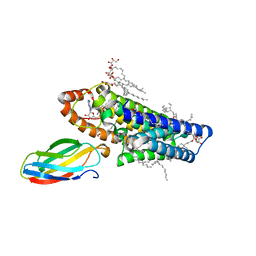 | | Atomic structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase), in complex with a monobody | | Descriptor: | DECANE, Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-03-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
5A43
 
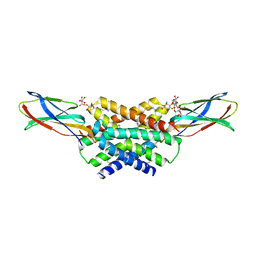 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, FLUORIDE ION, MONOBODIES, ... | | Authors: | Stockbridge, R.B, Kolmakova-Partensky, L, Shane, T, Koide, A, Koide, S, Miller, C, Newstead, S. | | Deposit date: | 2015-06-04 | | Release date: | 2015-09-02 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structures of a Double-Barrelled Fluoride Ion Channel.
Nature, 525, 2015
|
|
5A40
 
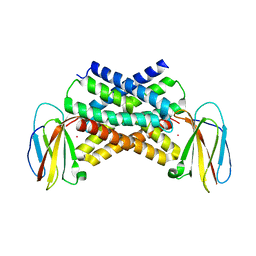 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | MERCURY (II) ION, MONOBODIES, PUTATIVE FLUORIDE ION TRANSPORTER CRCB | | Authors: | Stockbridge, R.B, Kolmakova-Partensky, L, Shane, T, Koide, A, Koide, S, Miller, C, Newstead, S. | | Deposit date: | 2015-06-04 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Double-Barrelled Fluoride Ion Channel.
Nature, 525, 2015
|
|
2R8S
 
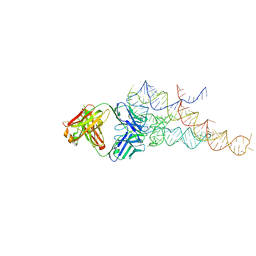 | | High resolution structure of a specific synthetic FAB bound to P4-P6 RNA ribozyme domain | | Descriptor: | Fab heavy chain, Fab light chain, MAGNESIUM ION, ... | | Authors: | Ye, J.D, Tereshko, V, Sidhu, S.S, Koide, S, Kossiakoff, A.A, Piccirilli, J.A. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthetic antibodies for specific recognition and crystallization of structured RNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5WOB
 
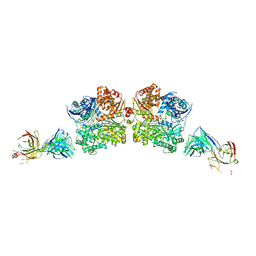 | | Crystal Structure Analysis of Fab1-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Insulin | | Descriptor: | IDE-bound Fab heavy chain, IDE-bound Fab light chain, Insulin, ... | | Authors: | McCord, L.A, Liang, W.G, Farcasanu, M, Wang, A.G, Koide, S, Tang, W.J. | | Deposit date: | 2017-08-01 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
7KZZ
 
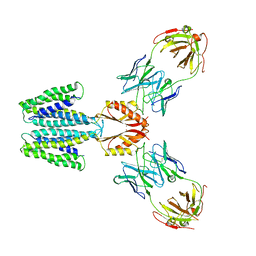 | | Cryo-EM structure of YiiP-Fab complex in Holo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain, ... | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
7KZX
 
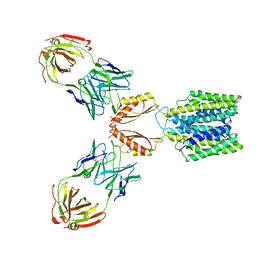 | | Cryo-EM structure of YiiP-Fab complex in Apo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
3RZW
 
 | |
7JW7
 
 | | Structure of monobody 27 human MLKL pseudokinase domain complex | | Descriptor: | Mixed lineage kinase domain-like protein, Monobody 27 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
7JXU
 
 | | Structure of monobody 32 human MLKL pseudokinase domain complex | | Descriptor: | 1,2-ETHANEDIOL, Mixed lineage kinase domain-like protein, Monobody 32 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
