2HID
 
 | |
4BVU
 
 | | Structure of Shigella effector OspG in complex with host UbcH5c- Ubiquitin conjugate | | Descriptor: | PROTEIN KINASE OSPG, UBIQUITIN, UBIQUITIN-CONJUGATING ENZYME E2 D3 | | Authors: | Pruneda, J.N, LeTrong, I, Stenkamp, R.E, Klevit, R.E, Brzovic, P.S. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | E2~Ub Conjugates Regulate the Kinase Activity of Shigella Effector Ospg During Pathogenesis.
Embo J., 33, 2014
|
|
2N0K
 
 | | Chemical shift assignments and structure of the alpha-crystallin domain from human, HSPB5 | | Descriptor: | Alpha-crystallin B chain | | Authors: | Rajagopal, P, Klevit, R.E, Shi, L, Baker, D. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis.
Elife, 4, 2015
|
|
3J07
 
 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3C5R
 
 | |
3FA2
 
 | |
3DBA
 
 | | Crystal structure of the cGMP-bound GAF a domain from the photoreceptor phosphodiesterase 6C | | Descriptor: | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha', GLYCEROL, GUANOSINE-3',5'-MONOPHOSPHATE | | Authors: | Martinez, S.E, Heikaus, C.C, Klevit, R.E, Beavo, J.A. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The Structure of the GAF A Domain from Phosphodiesterase 6C Reveals Determinants of cGMP Binding, a Conserved Binding Surface, and a Large cGMP-dependent Conformational Change.
J.Biol.Chem., 283, 2008
|
|
2KLR
 
 | | Solid-state NMR structure of the alpha-crystallin domain in alphaB-crystallin oligomers | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Rajagopal, P, Markovic, S, Bardiaux, B, Kuehne, R, Higman, V.A, Klevit, R.E, van Rossum, B, Oschkinat, H. | | Deposit date: | 2009-07-08 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4QPL
 
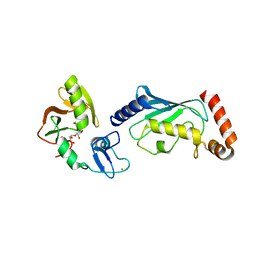 | | Crystal structure of RNF146(RING-WWE)/UbcH5a/iso-ADPr complex | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Wang, Z, DaRosa, P.A, Klevit, R.E, Xu, W. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal.
Nature, 517, 2015
|
|
3UGB
 
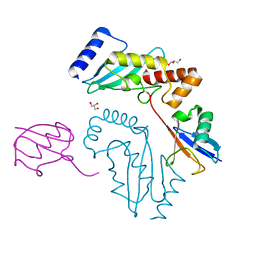 | | UbcH5c~Ubiquitin Conjugate | | Descriptor: | GLYCEROL, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Page, R.C, Pruneda, J.N, Klevit, R.E, Misra, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates.
Biochemistry, 51, 2012
|
|
6ALY
 
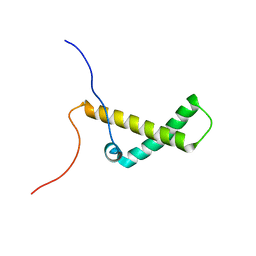 | | Solution structure of yeast Med15 ABD2 residues 277-368 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Tuttle, L.M, Pacheco, D, Warfield, L, Hahn, S, Klevit, R.E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex.
Cell Rep, 22, 2018
|
|
2FUH
 
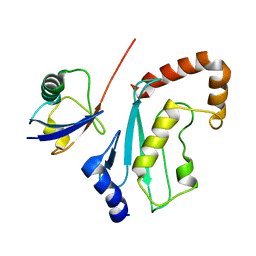 | | Solution Structure of the UbcH5c/Ub Non-covalent Complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Brzovic, P.S, Lissounov, A, Hoyt, D.W, Klevit, R.E. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A UbcH5/Ubiquitin Noncovalent Complex Is Required for Processive BRCA1-Directed Ubiquitination.
Mol.Cell, 21, 2006
|
|
1JEM
 
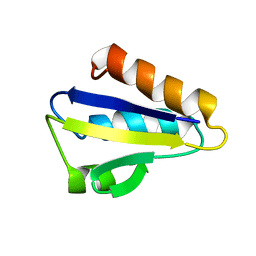 | | NMR STRUCTURE OF HISTIDINE PHOSPHORYLATED FORM OF THE PHOSPHOCARRIER HISTIDINE CONTAINING PROTEIN FROM BACILLUS SUBTILIS, NMR, 25 STRUCTURES | | Descriptor: | HISTIDINE CONTAINING PROTEIN | | Authors: | Jones, B.E, Rajagopal, P, Klevit, R.E. | | Deposit date: | 1997-04-01 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation on histidine is accompanied by localized structural changes in the phosphocarrier protein, HPr from Bacillus subtilis.
Protein Sci., 6, 1997
|
|
7JZV
 
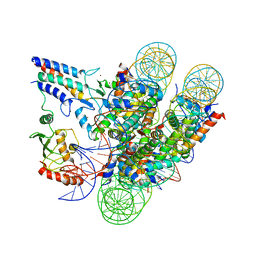 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6BP9
 
 | |
2LPB
 
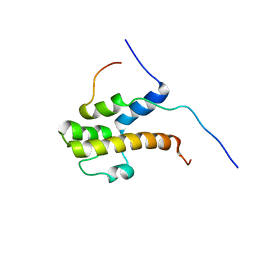 | | Structure of the complex of the central activation domain of Gcn4 bound to the mediator co-activator domain 1 of Gal11/med15 | | Descriptor: | General control protein GCN4, Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Brzovic, P.S, Heikaus, C.C, Kisselev, L, Vernon, R, Herbig, E, Pacheco, D, Warfield, L, Littlefield, P, Baker, D, Klevit, R.E, Hahn, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex.
Mol.Cell, 44, 2011
|
|
2MT6
 
 | | Solution structure of the human ubiquitin conjugating enzyme Ube2w | | Descriptor: | Ubiquitin-conjugating enzyme E2 W | | Authors: | Vittal, V, Shi, L, Wenzel, D.M, Brzovic, P.S, Klevit, R.E. | | Deposit date: | 2014-08-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intrinsic disorder drives N-terminal ubiquitination by Ube2w.
Nat.Chem.Biol., 11, 2015
|
|
2K31
 
 | | Solution Structure of cGMP-binding GAF domain of Phosphodiesterase 5 | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, Phosphodiesterase 5A, cGMP-specific | | Authors: | Heikaus, C.C, Stout, J.R, Sekharan, M.R, Eakin, C.M, Rajagopal, P, Brzovic, P.S, Beavo, J.A, Klevit, R.E. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the cGMP Binding GAF Domain from Phosphodiesterase 5: Insights into Nucleotide Selectivity, Dimerization, and cGMP-Dependent Conformational Change.
J.Biol.Chem., 283, 2008
|
|
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
1PAA
 
 | | STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Bernstein, B.E, Hoffman, R.C, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1994-07-15 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition.
Biochemistry, 33, 1994
|
|
1ARE
 
 | | STRUCTURES OF DNA-BINDING MUTANT ZINC FINGER DOMAINS: IMPLICATIONS FOR DNA BINDING | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Hoffman, R.C, Xu, R.X, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structures of DNA-binding mutant zinc finger domains: implications for DNA binding.
Protein Sci., 2, 1993
|
|
1ARF
 
 | | STRUCTURES OF DNA-BINDING MUTANT ZINC FINGER DOMAINS: IMPLICATIONS FOR DNA BINDING | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Hoffman, R.C, Xu, R.X, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structures of DNA-binding mutant zinc finger domains: implications for DNA binding.
Protein Sci., 2, 1993
|
|
1ARD
 
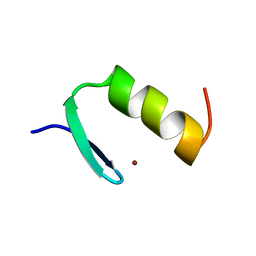 | | STRUCTURES OF DNA-BINDING MUTANT ZINC FINGER DOMAINS: IMPLICATIONS FOR DNA BINDING | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Hoffman, R.C, Xu, R.X, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structures of DNA-binding mutant zinc finger domains: implications for DNA binding.
Protein Sci., 2, 1993
|
|
2CSA
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor Basolateral Sorting Signal | | Descriptor: | Muscarinic acetylcholine receptor M3 | | Authors: | Iverson, H.A, Fox, D, Nadler, L.S, Klevit, R.E, Nathanson, N.M. | | Deposit date: | 2005-05-21 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Identification and structural determination of the M3 muscarinic acetylcholine receptor basolateral sorting signal.
J.Biol.Chem., 280, 2005
|
|
1JM7
 
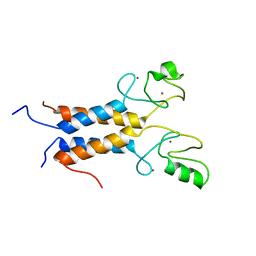 | | Solution structure of the BRCA1/BARD1 RING-domain heterodimer | | Descriptor: | BRCA1-ASSOCIATED RING DOMAIN PROTEIN 1, BREAST CANCER TYPE 1 SUSCEPTIBILITY PROTEIN, ZINC ION | | Authors: | Brzovic, P.S, Rajagopal, P, Hoyt, D.W, King, M.-C, Klevit, R.E. | | Deposit date: | 2001-07-17 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a BRCA1-BARD1 heterodimeric RING-RING complex.
Nat.Struct.Biol., 8, 2001
|
|
