6Y4S
 
 | |
6SJU
 
 | |
6SHI
 
 | |
6SHH
 
 | |
5WPI
 
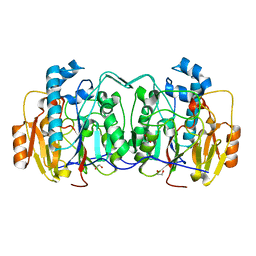 | |
5KW9
 
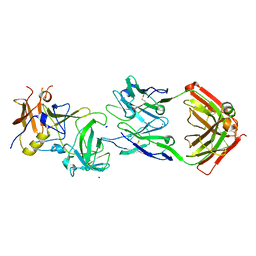 | |
3SLN
 
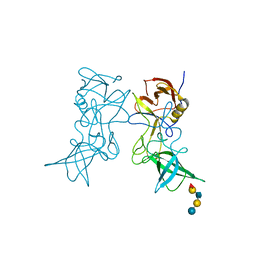 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to H pentasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SEJ
 
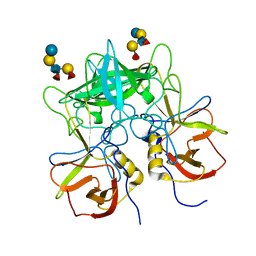 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to Secretor Lewis HBGA (LeB) | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SLD
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to A trisaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SKB
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SJP
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid, ZINC ION | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
4P26
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with A-type 2 HBGA | | Descriptor: | P domain of VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P1V
 
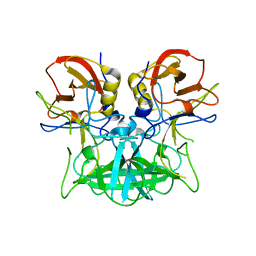 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P3I
 
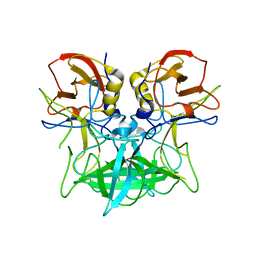 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P12
 
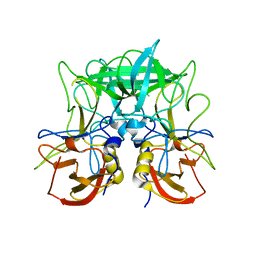 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | Descriptor: | Major capsid protein | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6011 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P25
 
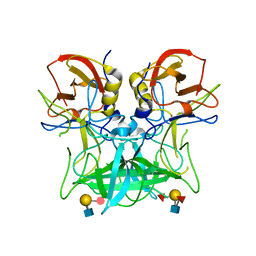 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4991 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P2N
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeX HBGA | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
3JYI
 
 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
4IMZ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN1
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, SULFATE ION | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMQ
 
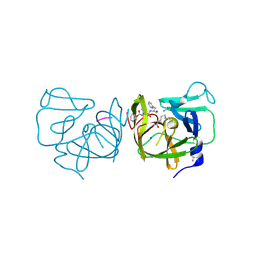 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN2
 
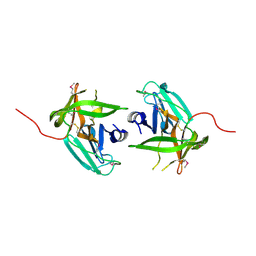 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
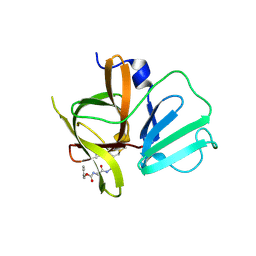 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4F6H
 
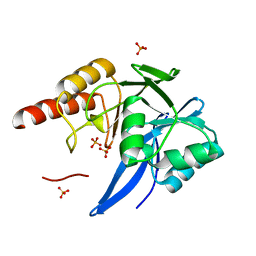 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
4F6Z
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, CITRATE ANION, ZINC ION | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-15 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
