2L97
 
 | |
2L83
 
 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
5F9P
 
 | | Crystal structure study of anthrone oxidase-like protein | | Descriptor: | Anthrone oxidase-like protein, GLYCEROL | | Authors: | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
2LJI
 
 | |
2KI0
 
 | | NMR Structure of a de novo designed beta alpha beta | | Descriptor: | DS119 | | Authors: | Liang, H, Chen, H, Fan, K, Wei, P, Guo, X, Jin, C, Zeng, C, Tang, C, Lai, L. | | Deposit date: | 2009-04-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | De novo design of a beta alpha beta motif.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7CK8
 
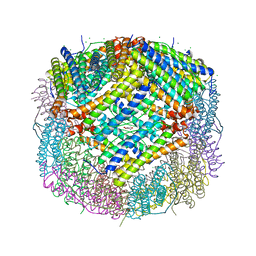 | | Crystal structure of human ferritin heavy chain mutant C90S/C102S/C130S | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Chen, X, Jiang, B, Yan, X, Fan, K. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A natural drug entry channel in the ferritin nanocage.
Nano Today, 35, 2020
|
|
7CK9
 
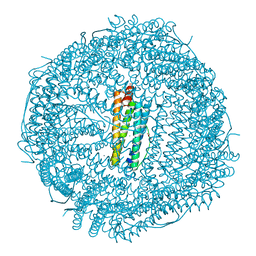 | | Crystal structure of Doxorubicin loaded human ferritin heavy chain | | Descriptor: | CHLORIDE ION, Ferritin heavy chain, GLYCEROL, ... | | Authors: | Chen, X, Jiang, B, Yan, X, Fan, K. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | A natural drug entry channel in the ferritin nanocage.
Nano Today, 35, 2020
|
|
1UHL
 
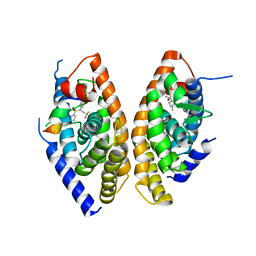 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
6GZY
 
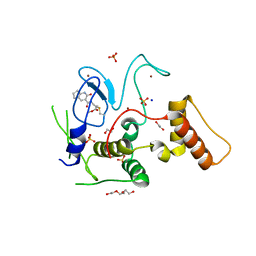 | | HOIP-fragment5 complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF31, SODIUM ION, ... | | Authors: | Johansson, H, Tsai, Y.C.I, Fantom, K, Chung, C.W, Martino, L, House, D, Rittinger, K. | | Deposit date: | 2018-07-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Covalent Ligand Screening Enables Rapid Discovery of Inhibitors for the RBR E3 Ubiquitin Ligase HOIP.
J. Am. Chem. Soc., 141, 2019
|
|
1XSE
 
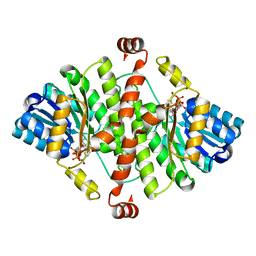 | | Crystal Structure of Guinea Pig 11beta-Hydroxysteroid Dehydrogenase Type 1 | | Descriptor: | 11beta-hydroxysteroid dehydrogenase type 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogg, D, Elleby, B, Norstrom, C, Stefansson, K, Abrahmsen, L, Oppermann, U, Svensson, S. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guinea pig 11beta-hydroxysteroid dehydrogenase type 1 provides a model for enzyme-lipid bilayer interactions
J.Biol.Chem., 280, 2005
|
|
5VLR
 
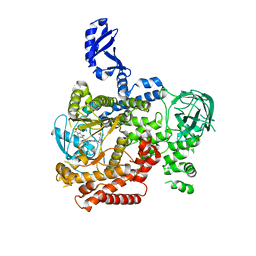 | | CRYSTAL STRUCTURE OF PI3K DELTA IN COMPLEX WITH A TRIFLUORO-ETHYL-PYRAZOL-PYROLOTRIAZINE INHIBITOR | | Descriptor: | 4-acetyl-1-(3-{4-amino-5-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)-3,3-dimethylpiperazin-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Sack, J.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Potent, Selective, and Efficacious Phosphatidylinositol 3-Kinase delta (PI3K delta ) Inhibitor for the Treatment of Immunological Disorders.
J. Med. Chem., 60, 2017
|
|
2L32
 
 | |
3OOZ
 
 | | Bace1 in complex with the aminohydantoin Compound 102 | | Descriptor: | (5R)-2-amino-5-[4-(difluoromethoxy)phenyl]-5-[4-fluoro-3-(5-fluoropent-1-yn-1-yl)phenyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Aminohydantoins as Potent and Selective Human beta-Secretase (BACE1) Inhibitors with Enhanced Brain Permeability
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3IGB
 
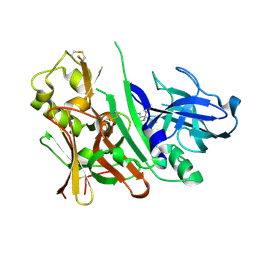 | | Bace-1 with Compound 3 | | Descriptor: | 8,8-diphenyl-2,3,4,8-tetrahydroimidazo[1,5-a]pyrimidin-6-amine, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Aminoimidazoles as potent and selective human beta-secretase (BACE1) inhibitors
J.Med.Chem., 52, 2009
|
|
3IN4
 
 | | Bace1 with Compound 38 | | Descriptor: | (5S)-2-amino-5-(2,6-diethylpyridin-4-yl)-3-methyl-5-(3-pyrimidin-5-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
3IN3
 
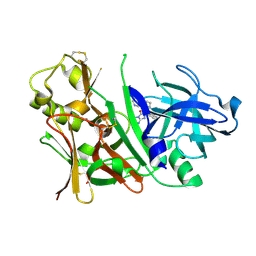 | | Bace1 with Compound 30 | | Descriptor: | (5S)-2-amino-3-methyl-5-pyridin-4-yl-5-(3-pyridin-3-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
3L38
 
 | | Bace1 in complex with the aminopyridine Compound 44 | | Descriptor: | 6-({2-(2-chlorophenyl)-5-[4-(pyrimidin-5-yloxy)phenyl]-1H-pyrrol-1-yl}methyl)pyridin-2-amine, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L3A
 
 | | Bace-1 with the aminopyridine Compound 32 | | Descriptor: | 4-(4-{1-[(6-aminopyridin-2-yl)methyl]-5-(2-chlorophenyl)-1H-pyrrol-2-yl}phenoxy)butanenitrile, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8GBU
 
 | | Hepatitis B capsid Y132A mutant with compound AB-506 | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(1S)-4-[(3-chloro-4-fluorophenyl)carbamoyl]-7-fluoro-2,3-dihydro-1H-inden-1-yl}carbamate, 1,2-ETHANEDIOL, Capsid protein | | Authors: | Horanyi, P.S, Mayclin, S.J. | | Deposit date: | 2023-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and structure-activity relationship of a bicyclic HBV capsid assembly modulator chemotype leading to the identification of clinical candidate AB-506.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
3MTS
 
 | | Chromo Domain of Human Histone-Lysine N-Methyltransferase SUV39H1 | | Descriptor: | Histone-lysine N-methyltransferase SUV39H1 | | Authors: | Lam, R, Li, Z, Wang, J, Crombet, L, Walker, J.R, Ouyang, H, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human SUV39H1 Chromodomain and Its Recognition of Histone H3K9me2/3.
Plos One, 7, 2012
|
|
6KU8
 
 | | structure of HRV-C 3C protein with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Genome polyprotein | | Authors: | Zhu, L, Yuan, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design.
Virol Sin, 35, 2020
|
|
6KU7
 
 | | structure of HRV-C 3C protein | | Descriptor: | Genome polyprotein | | Authors: | Zhu, L, Yuan, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design.
Virol Sin, 35, 2020
|
|
6J0Z
 
 | | Crystal structure of AlpK | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative angucycline-like polyketide oxygenase | | Authors: | Wang, W, Liu, Y, Liang, H. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Crystal structure of AlpK: An essential monooxygenase involved in the biosynthesis of kinamycin
Biochem. Biophys. Res. Commun., 510, 2019
|
|
