5ODN
 
 | |
5OQO
 
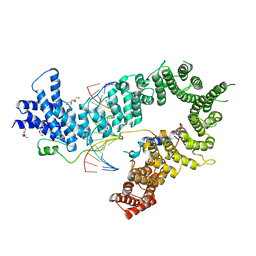 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Condensin complex subunit 2, Condensin complex subunit 3, ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
5OQN
 
 | |
5OQQ
 
 | |
5OQP
 
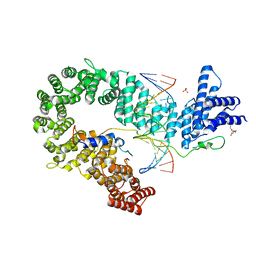 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form I) | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, DNA (5'-D(*GP*AP*TP*GP*TP*GP*TP*AP*GP*CP*TP*AP*CP*AP*CP*AP*TP*C)-3'), ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
5OQR
 
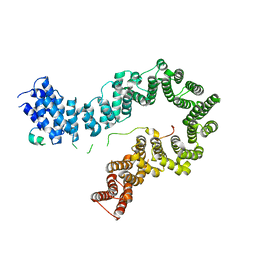 | |
6ZTW
 
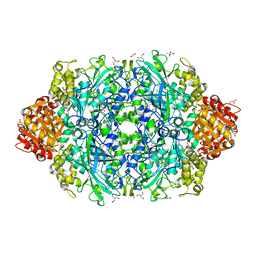 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
4TX0
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-(2-methoxyethoxy)-3-(2-methoxyethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-(2-methoxyethoxy)-3-(2-methoxyethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bischoff, M, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Stereoselective Construction of the 5-Hydroxy Diazabicyclo[4.3.1]decane-2-one Scaffold, a Privileged Motif for FK506-Binding Proteins.
Org.Lett., 16, 2014
|
|
5ZNF
 
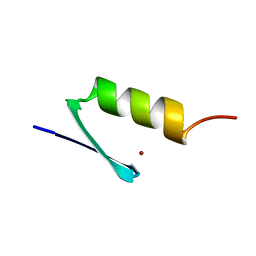 | |
8C58
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-hydroxycytosine and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5OC)P*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C59
 
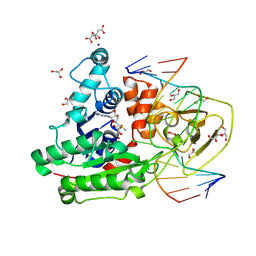 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-bromocytosine (converted to 5mC) and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, CITRIC ACID, Cytosine-specific methyltransferase, ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C57
 
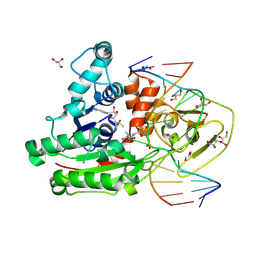 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5,6-dihydro-5-azacytosine (converted to 5m-dhaC) and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5MA)P*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C56
 
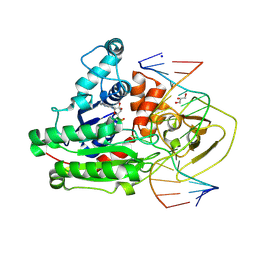 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 2'-deoxy-5-methylzebularine (5mZ) and 5-methylcytosine containing dsDNA | | Descriptor: | Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5PY)P*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8F0Q
 
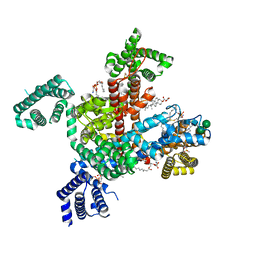 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-cyclopropyl-4-({1-[(1S)-1-(3,5-dichlorophenyl)ethyl]piperidin-4-yl}methoxy)-2-fluoro-N-(methanesulfonyl)benzamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0S
 
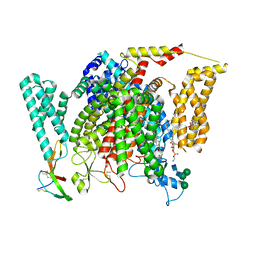 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0P
 
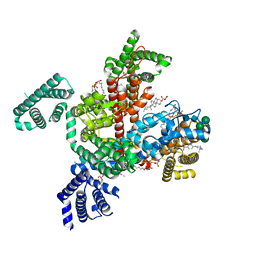 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0R
 
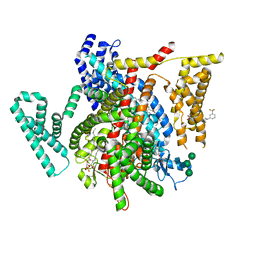 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
5LS0
 
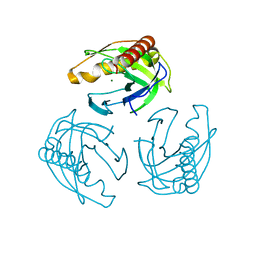 | | Crystal structure of Inorganic Pyrophosphatase PPA1 from Arabidopsis thaliana | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Soluble inorganic pyrophosphatase 1 | | Authors: | Grzechowiak, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity.
Biochem.J., 476, 2019
|
|
5ODP
 
 | |
6ZTX
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6ZTV
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
8BQ1
 
 | | Herpes simplex virus type 1 protease | | Descriptor: | Assemblin, GLYCEROL | | Authors: | Pachota, M, Grzywa, R, Plewka, J, Wilk, P, Mackereth, C, Czarna, A, Sienczyk, M, Pyrc, K. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Herpes simplex virus type 1 protease
To Be Published
|
|
7SX3
 
 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 1/2 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
7SX4
 
 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 2/2 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
6XIW
 
 | | Cryo-EM structure of the sodium leak channel NALCN-FAM155A complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Noland, C.L, Weidling, C, Clairfeuille, T, Bahlke, O.O, Ameen, A.O, Li, Z.R, Arthur, C.P, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human sodium leak channel NALCN.
Nature, 587, 2020
|
|
