1PNJ
 
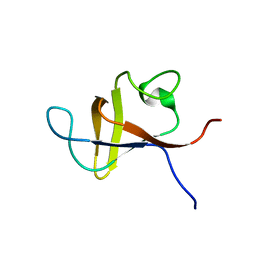 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
2PNB
 
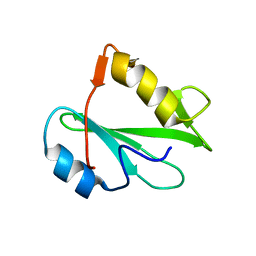 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
2PNI
 
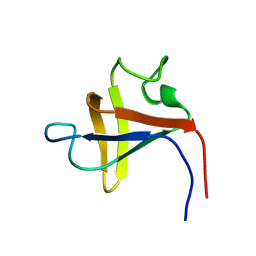 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
2PNA
 
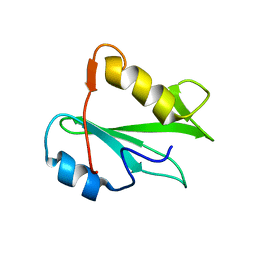 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
1C8P
 
 | | NMR STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE COMMON BETA-CHAIN IN THE GM-CSF, IL-3 AND IL-5 RECEPTORS | | Descriptor: | CYTOKINE RECEPTOR COMMON BETA CHAIN | | Authors: | Mulhern, T.D, D'Andrea, R.J, Gaunt, C, Vandeleur, L, Vadas, M.A, Lopez, A.F, Booker, G.W, Bagley, C.J. | | Deposit date: | 1999-10-05 | | Release date: | 2000-06-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the cytokine-binding domain of the common beta-chain of the receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3 and interleukin-5.
J.Mol.Biol., 297, 2000
|
|
4WOP
 
 | | Nucleotide Triphosphate Promiscuity in Mycobacterium tuberculosis Dethiobiotin Synthetase | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, SULFATE ION | | Authors: | Salaemae, W, Yap, M.Y, Wegener, K.L, Booker, G.W, Wilce, M.C.J, Polyak, S.W. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Nucleotide triphosphate promiscuity in Mycobacterium tuberculosis dethiobiotin synthetase.
Tuberculosis (Edinb), 95, 2015
|
|
1GL5
 
 | |
1WLX
 
 | |
1WR4
 
 | |
1WR7
 
 | |
1WMV
 
 | |
1WR3
 
 | |
4DQ2
 
 | | Structure of staphylococcus aureus biotin protein ligase in complex with biotinol-5'-amp | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Wilce, M, Yap, M, Pendini, N, Soares de Costa, T, Polyak, S, Tieu, W, Booker, G, Wallace, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
6CZD
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8ENI
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 3-[4-(5-fluoro-4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-5-methyl-1,3-benzoxazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Wilce, M.C.J, Cini, D.A. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
7JT6
 
 | | Mycobacterium tuberculosis dethiobiotin synthetase in complex with Tetrazole 2 | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, GLYCEROL, SULFATE ION, ... | | Authors: | Pederick, J.P, Bean, J.H, Bruning, J.B. | | Deposit date: | 2020-08-17 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Dethiobiotin Synthase ( Mt DTBS): Toward Next-Generation Antituberculosis Agents.
Acs Chem.Biol., 16, 2021
|
|
7JT5
 
 | |
7L1J
 
 | |
6APW
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 4-[(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)methyl]benzoic acid, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
6AQQ
 
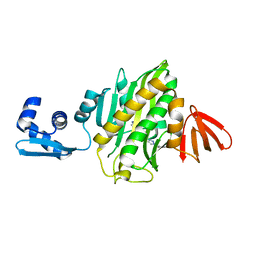 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | (3aS,4S,6aR)-4-(5-{1-[(3-fluorophenyl)methyl]-1H-1,2,3-triazol-4-yl}pentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
6NDL
 
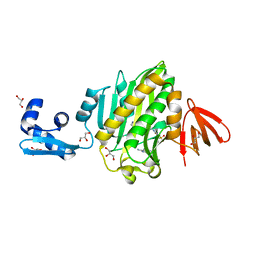 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
6CVF
 
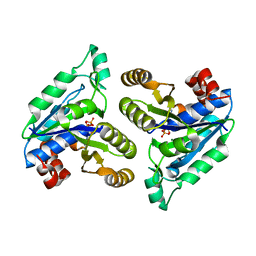 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine diphosphate | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Bruning, J.B, Wegener, K.L, Polyak, S.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CZB
 
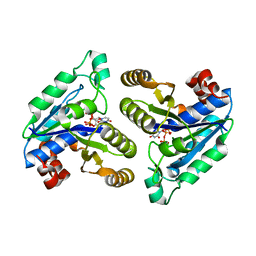 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with uridine triphosphate (UTP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CVE
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin Synthetase in complex with cytidine triphosphate and 7,8-diaminopelargonic acid | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ATP-dependent dethiobiotin synthetase BioD, CITRATE ANION, ... | | Authors: | Thompson, A.P, Bruning, J.B, Wegener, K.L, Polyak, S.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CVV
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with adenosine triphosphate (ATP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-03-29 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
