2RAH
 
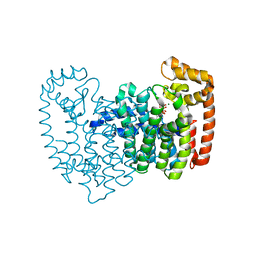 | | Human FDPS synthase in complex with novel inhibitor | | Descriptor: | Farnesyl pyrophosphate synthetase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Evdokimov, A.G, Barnett, B.L, Ebetino, F.H, Pokross, M. | | Deposit date: | 2007-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human FDPS synthase in complex with novel inhibitor
To be Published
|
|
4WA6
 
 | |
2R59
 
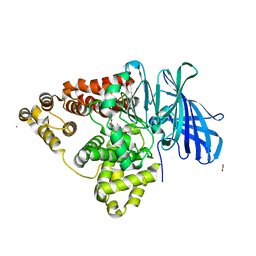 | | Leukotriene A4 hydrolase complexed with inhibitor RB3041 | | Descriptor: | ACETIC ACID, Leukotriene A-4 hydrolase, N-{(2S)-3-[(R)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Tholander, F, Haeggstrom, J.Z, Thunnissen, M, Muroya, A, Roques, B.P, Fournie-Zaluski, M.C. | | Deposit date: | 2007-09-03 | | Release date: | 2008-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
2REO
 
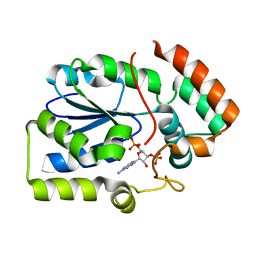 | | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Putative sulfotransferase 1C3 | | Authors: | Tempel, W, Pan, P, Dong, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP.
To be Published
|
|
2REY
 
 | | Crystal structure of the PDZ domain of human dishevelled 2 (homologous to Drosophila dsh) | | Descriptor: | Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Papagrigoriou, E, Gileadi, C, Elkins, J, Cooper, C, Ugochukwu, E, Turnbull, A, Pike, A.C.W, Gileadi, O, von Delft, F, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-27 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the PDZ domains of human dishevelled 2 (homologous to Drosophila dsh).
To be Published
|
|
2RF3
 
 | | Crystal Structure of Tricyclo-DNA: An Unusual Compensatory Change of Two Adjacent Backbone Torsion Angles | | Descriptor: | 5'-d(CGCG(TCY)ATTCGCG)-3', SPERMINE, ZINC ION | | Authors: | Pallan, P.S, Ittig, D, Heroux, A, Wawrzak, Z, Leumann, C.J, Egli, M. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles.
Chem.Commun.(Camb.), 2008, 2008
|
|
4WD6
 
 | |
2R5R
 
 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
4UVP
 
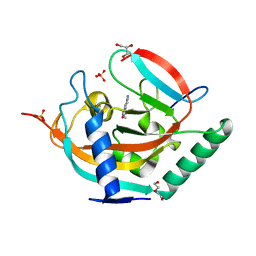 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- ethyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-ethylisoquinolin-1(2H)-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4V59
 
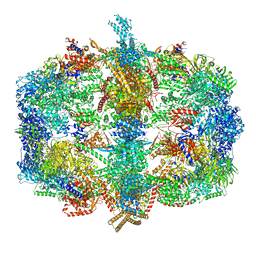 | | Crystal structure of fatty acid synthase complexed with nadp+ from thermomyces lanuginosus at 3.1 angstrom resolution. | | Descriptor: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | Deposit date: | 2007-03-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
4UWY
 
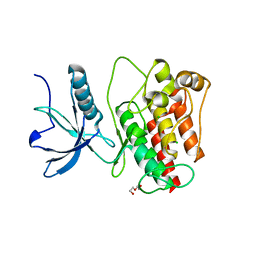 | | FGFR1 Apo structure | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Thiyagarajan, N, Bunney, T, Katan, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
2RA2
 
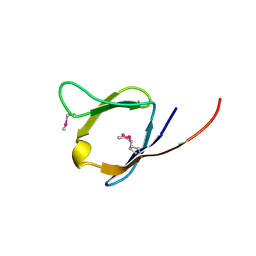 | | X-Ray structure of the Q7CPV8 protein from Salmonella typhimurium at the resolution 1.9 A. Northeast Structural Genomics Consortium target StR88A | | Descriptor: | Putative lipoprotein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Vorobiev, S.M, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray structure of the Q7CPV8 protein from Salmonella typhimurium at the resolution 1.9 A.
To be Published
|
|
4UYR
 
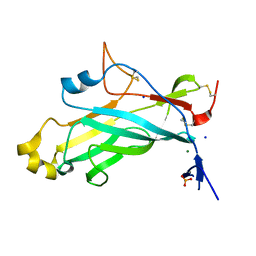 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Veelders, M, Kraushaar, T, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4UZ6
 
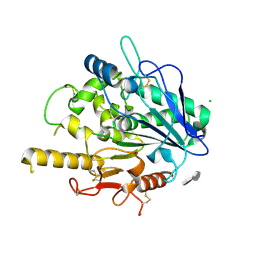 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM V - SOS COMPLEX - 1.9A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
2R8I
 
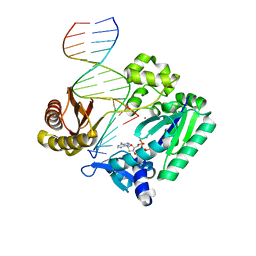 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
4V4S
 
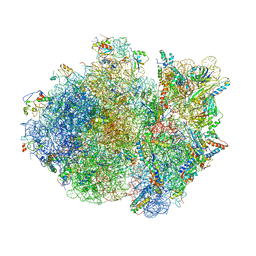 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V62
 
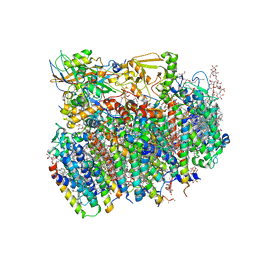 | | Crystal Structure of cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Guskov, A, Gabdulkhakov, A, Kern, J, Broser, M, Zouni, A, Saenger, W. | | Deposit date: | 2008-01-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride
Nat.Struct.Mol.Biol., 16, 2009
|
|
4V8B
 
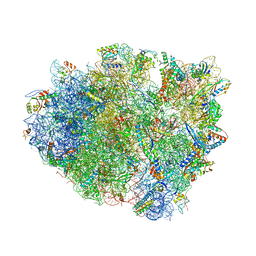 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-06 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4W4N
 
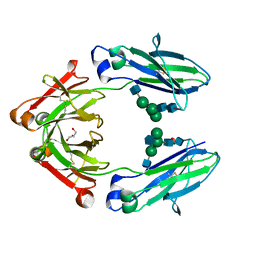 | | Crystal structure of human Fc at 1.80 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
2RCU
 
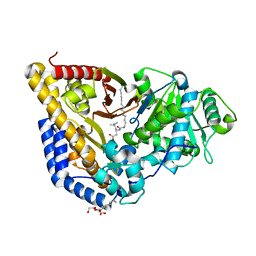 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with r-3-(hexadecanoylamino)-4-(trimethylazaniumyl)butanoate | | Descriptor: | (3R)-3-(hexadecanoylamino)-4-(trimethylammonio)butanoate, Carnitine O-palmitoyltransferase 2, octyl beta-D-glucopyranoside | | Authors: | Rufer, A.C, Benz, J, Chomienne, O, Thoma, R, Hennig, M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Carnitine palmitoyltransferase 2: analysis of membrane association and complex structure with a substrate analog.
Febs Lett., 581, 2007
|
|
4V6F
 
 | | Elongation complex of the 70S ribosome with three tRNAs and mRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 23S RRNA, ... | | Authors: | Jenner, L.B, Yusupova, G, Yusupov, M. | | Deposit date: | 2009-07-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural aspects of messenger RNA reading frame maintenance by the ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2R7P
 
 | | Crystal Structure of H225A NSP2 and AMPPNP complex | | Descriptor: | Non-structural RNA-binding protein 35, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kumar, M, Prasad, B.V.V. | | Deposit date: | 2007-09-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and Biochemical Analysis of Rotavirus NSP2 with Nucleotides Reveals a Nucleoside Diphosphate Kinase-Like Activity
J.Virol., 81, 2007
|
|
1W3Z
 
 | | SeMet derivative of BbCRASP-1 from Borrelia Burgdorferi | | Descriptor: | BBCRASP-1 | | Authors: | Cordes, F.S, Roversi, P, Goodstadt, L, Ponting, C, Kraiczy, P, Skerka, C, Kirschfink, M, Simon, M.M, Brade, V, Zipfel, P, Wallich, R, Lea, S.M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Novel Fold for the Factor H-Binding Protein Bbcrasp-1 of Borrelia Burgdorferi
Nat.Struct.Mol.Biol., 12, 2005
|
|
2R8H
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
