7YVJ
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH236 Fab | | Descriptor: | Spike glycoprotein, TH236 Fab heavy chain, TH236 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVL
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH272 Fab heavy chain, TH272 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVI
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH236 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
6SDG
 
 | |
7WQ4
 
 | | Galanin-bound galanin receptor 2 in complex with Gq | | Descriptor: | CHOLESTEROL, Engineered Guanine nucleotide-binding protein G(q) subunit alpha, Galanin, ... | | Authors: | Duan, J, Shen, D.D, Xu, H.E, Zhang, Y, Jiang, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis for allosteric agonism and G protein subtype selectivity of galanin receptors
Nat Commun, 13, 2022
|
|
7WQ3
 
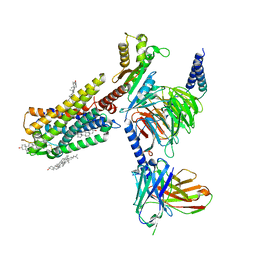 | | Galanin-bound galanin receptor 1 in complex with Gi | | Descriptor: | CHOLESTEROL, Galanin, Galanin receptor type 1, ... | | Authors: | Duan, J, Shen, D.D, Xu, H.E, Zhang, Y, Jiang, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for allosteric agonism and G protein subtype selectivity of galanin receptors
Nat Commun, 13, 2022
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6XAA
 
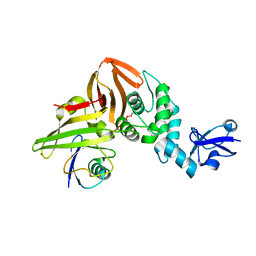 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
4PSB
 
 | |
7DRE
 
 | | Cryo-EM structure of DfgA-B at 2.54 angstrom resolution | | Descriptor: | DfgB, Sugar phosphate isomerase/epimerase | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRD
 
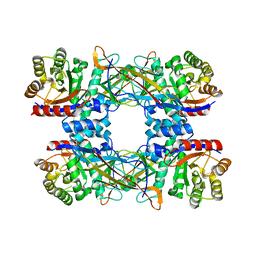 | | Cryo-EM structure of DgpB-C at 2.85 angstrom resolution | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7WWF
 
 | | Crystal structure of BioH3 from Mycolicibacterium smegmatis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Esterase | | Authors: | Yang, J, Xu, Y.C, Gan, J.H, Feng, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-06 | | Last modified: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Three enigmatic BioH isoenzymes are programmed in the early stage of mycobacterial biotin synthesis, an attractive anti-TB drug target.
Plos Pathog., 18, 2022
|
|
7XN2
 
 | | Crystal structure of CvkR, a novel MerR-type transcriptional regulator | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alr3614 protein, DI(HYDROXYETHYL)ETHER | | Authors: | Liang, Y.J, Zhu, T, Ma, H.L, Lu, X.F, Hess, W.R. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CvkR is a MerR-type transcriptional repressor of class 2 type V-K CRISPR-associated transposase systems.
Nat Commun, 14, 2023
|
|
7WVD
 
 | | Crystal structure of H14 complexed with SIA28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SIA28, ... | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7WVI
 
 | | Crystal structure of SIA28 | | Descriptor: | Heavy chain of SIA28, Light chain of SIA28 | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7WVG
 
 | | Crystal structure of H18 complexed with SIA28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SIA28, ... | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
1UL7
 
 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kinase-associated domain 1 of mouse microtubule-associated protein/microtubule affinity-regulating kinase 3
Protein Sci., 15, 2006
|
|
7VMU
 
 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | Spike protein S1, scFv E4 | | Authors: | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
2YT0
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2YT1
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
3VQJ
 
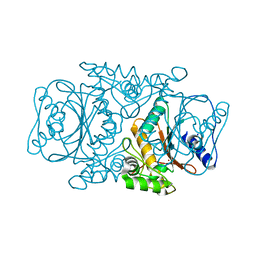 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, ZINC ION | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-03-24 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VRK
 
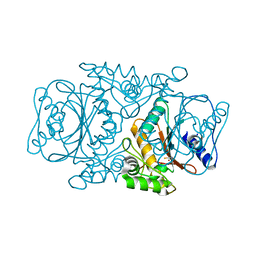 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-04-11 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
5B3T
 
 | |
2RR7
 
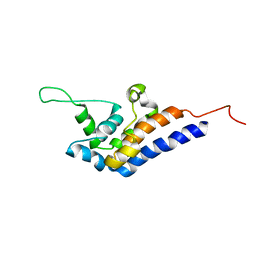 | | Microtubule Binding Domain of DYNEIN-C | | Descriptor: | Dynein heavy chain 9 | | Authors: | Kato, Y, Yagi, T, Ohki, S, Burgess, S, Honda, S, Kamiya, R, Tanokura, M. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the microtubule-binding domain of flagellar dynein
Structure, 22, 2014
|
|
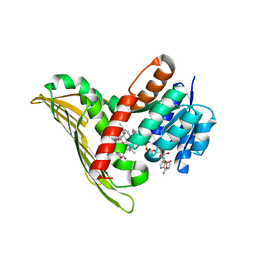
5B3V
 
 | |
