1T2M
 
 | | Solution Structure Of The Pdz Domain Of AF-6 | | Descriptor: | AF-6 protein | | Authors: | Zhou, H, Wu, J.H, Xu, Y.Q, Huang, A.D, Shi, Y.Y. | | Deposit date: | 2004-04-22 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of AF-6 PDZ Domain and Its Interaction with the C-terminal Peptides from Neurexin and Bcr
J.Biol.Chem., 280, 2005
|
|
4PPH
 
 | | Crystal structure of conglutin gamma, a unique basic 7S globulin from lupine seeds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Czubinski, J, Barciszewski, J, Gilski, M, Lampart-Szczapa, E, Jaskolski, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structure of gamma-conglutin: insight into the quaternary structure of 7S basic globulins from legumes.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VQT
 
 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3VR1
 
 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-04-03 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3W69
 
 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | (5R,6S)-2-[((2S,5R)-2-{[(3R)-4-acetyl-3-methylpiperazin-1-yl]carbonyl}-5-ethylpyrrolidin-1-yl)carbonyl]-5,6-bis(4-chlorophenyl)-3-isopropyl-6-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of novel orally active p53-MDM2 interaction inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
7EIU
 
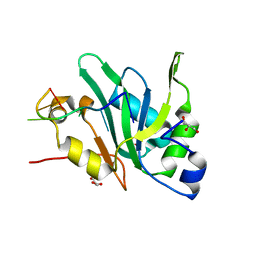 | | Crystal structure of Mei2 RRM3 in complex with 8mer meiRNA | | Descriptor: | GLYCEROL, Meiosis protein mei2, RNA (5'-R(P*UP*UP*CP*UP*GP*C)-3') | | Authors: | Shen, S.Y, Lv, M.Q. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
7EIO
 
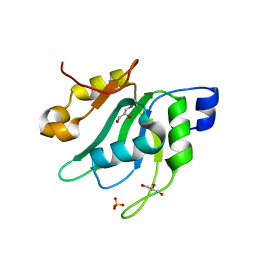 | | Crystal Structure of Mei2 RRM3 | | Descriptor: | GLYCEROL, Meiosis protein mei2, SULFATE ION | | Authors: | Shen, S.Y, Li, F.D. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
7E1Z
 
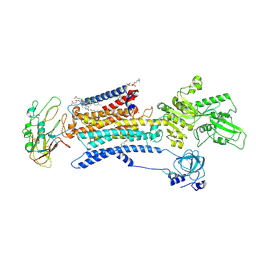 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E20
 
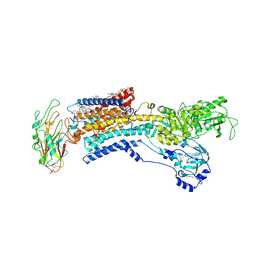 | | Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E21
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7DKJ
 
 | |
1FXI
 
 | |
7VNE
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113.1 (UUU-state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VND
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UUD-state, state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNB
 
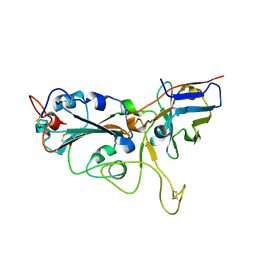 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNC
 
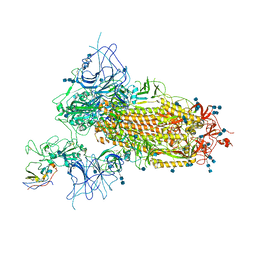 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
7DNN
 
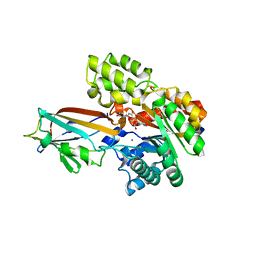 | | Crystal structure of the AgCarB2-C2 complex with homoorientin | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-6-[(2S,3R,4R,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-5,7-bis(oxidanyl)chromen-4-one, AP_endonuc_2 domain-containing protein, AgCarC2, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DNM
 
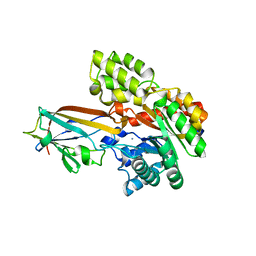 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
6FEK
 
 | |
7DTK
 
 | |
7DTJ
 
 | |
1HSR
 
 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
2ROM
 
 | |
8A9I
 
 | | PI3KC2a core in complex with PITCOIN1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-~{N}-(1,3-thiazol-2-yl)ethanamide, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
