3GKJ
 
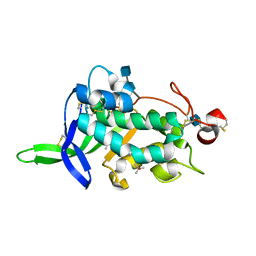 | | NPC1D(NTD):25hydroxycholesterol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 25-HYDROXYCHOLESTEROL, ... | | Authors: | Kwon, H.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GKI
 
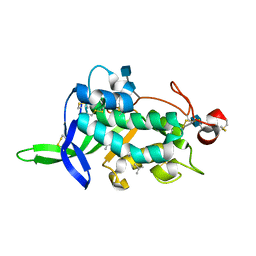 | | NPC1(NTD):cholesterol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Kwon, H.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol.
Cell(Cambridge,Mass.), 137, 2009
|
|
6U7E
 
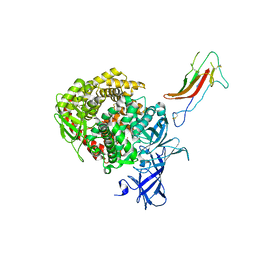 | | HCoV-229E RBD Class III in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7G
 
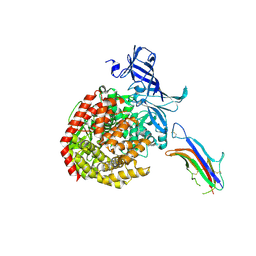 | | HCoV-229E RBD Class V in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7F
 
 | | HCoV-229E RBD Class IV in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
9ETZ
 
 | | III2IV respiratory supercomplex from Saccharomyces cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CALCIUM ION, ... | | Authors: | Moe, A, Brzezinski, P. | | Deposit date: | 2024-03-27 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Electron transfer in the respiratory chain at low salinity.
Nat Commun, 15, 2024
|
|
6X87
 
 | | CryoEM structure of the Plasmodium berghei circumsporozoite protein in complex with inhibitory mouse antibody 3D11. | | Descriptor: | 3D11 Fab heavy chain, 3D11 Fab kappa chain, Circumsporozoite protein | | Authors: | Kucharska, I, Thai, E, Rubinstein, J, Julien, J.P. | | Deposit date: | 2020-06-01 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural ordering of the Plasmodium berghei circumsporozoite protein repeats by inhibitory antibody 3D11.
Elife, 9, 2020
|
|
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
8DNN
 
 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
1A1X
 
 | | CRYSTAL STRUCTURE OF MTCP-1 INVOLVED IN T CELL MALIGNANCIES | | Descriptor: | HMTCP-1 | | Authors: | Fu, Z.Q, Dubois, G.C, Song, S.P, Kulikovskaya, I, Virgilio, L, Rothstein, J, Croce, C.M, Weber, I.T, Harrison, R.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6X8P
 
 | |
6X8Q
 
 | |
6X8U
 
 | |
7RLZ
 
 | |
7RM1
 
 | |
7RLX
 
 | |
7RLY
 
 | |
7RLV
 
 | |
7RM0
 
 | |
7RLW
 
 | |
7RM3
 
 | | Antibody 2E10.E9 in complex with P. vivax CSP peptide ANGAGNQPGANGAGNQPGANGAGGQAA | | Descriptor: | 2E10.E9 Fab heavy chain, 2E10.E9 Fab light chain, ACETATE ION, ... | | Authors: | Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of Plasmodium vivax inhibition by antibodies binding to the circumsporozoite protein repeats.
Elife, 11, 2022
|
|
6X8S
 
 | |
5UF5
 
 | | Structure of the effector protein SidK (lpg0968) from Legionella pneumophila (domain-swapped dimer) | | Descriptor: | GLYCEROL, effector protein SidK | | Authors: | Beyrakhova, K, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5VL3
 
 | | CD22 d1-d3 in complex with therapeutic Fab Epratuzumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell receptor CD22, Epratuzumab Fab Heavy Chain, ... | | Authors: | Sicard, T, Ereno-Orbea, J, Julien, J.P. | | Deposit date: | 2017-04-24 | | Release date: | 2017-10-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
3B8O
 
 | |
