8JDY
 
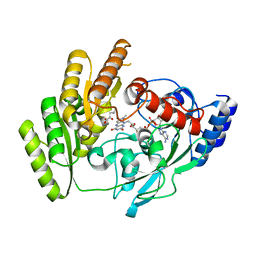 | | Crystal structure of mLDHD in complex with 2-ketoisocaproic acid | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDD
 
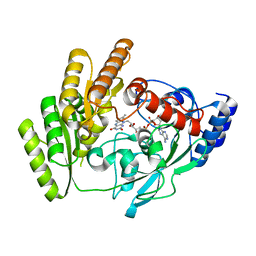 | | Crystal structure of H405A mLDHD in apo form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, mitochondrial | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDZ
 
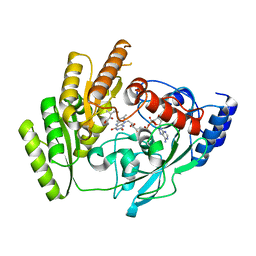 | | Crystal structure of mLDHD in complex with 2-keto-3-methylvaleric acid | | Descriptor: | (3S)-3-methyl-2-oxopentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDF
 
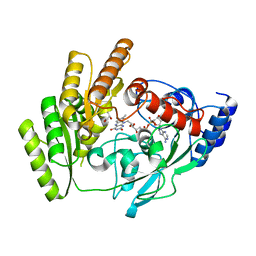 | | Crystal structure of H405A mLDHD in complex with D-lactate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDX
 
 | | Crystal structure of mLDHD in complex with 2-ketoisovaleric acid | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDE
 
 | | Crystal structure of mLDHD in complex with D-lactate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDO
 
 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyhexanoic acid | | Descriptor: | (2R)-2-hydroxyhexanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDP
 
 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyisovaleric acid | | Descriptor: | DEAMINOHYDROXYVALINE, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
7D0G
 
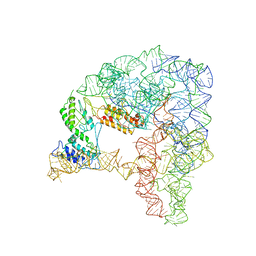 | | Cryo-EM structure of a pre-catalytic group II intron | | Descriptor: | Group II intron-encoded protein LtrA, RNA (714-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7D1A
 
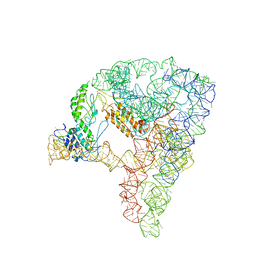 | | cryo-EM structure of a group II intron RNP complexed with its reverse transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3'), RNA (692-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
3NWN
 
 | | Crystal structure of the human KIF9 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF9, ... | | Authors: | Zhu, H, Tempel, W, He, H, Shen, Y, Wang, J, Brothers, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human KIF9 motor domain in complex with ADP
To be Published
|
|
3K6U
 
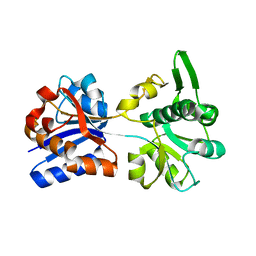 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5IYC
 
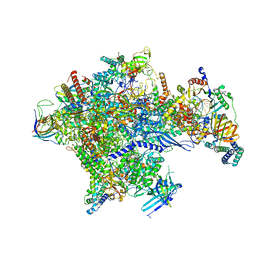 | | Human core-PIC in the initial transcribing state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
8SDU
 
 | | Structure of rat organic anion transporter 1 (OAT1) | | Descriptor: | Solute carrier family 22 member 6 | | Authors: | Dou, T, Jiang, J. | | Deposit date: | 2023-04-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | The substrate and inhibitor binding mechanism of polyspecific transporter OAT1 revealed by high-resolution cryo-EM.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SDZ
 
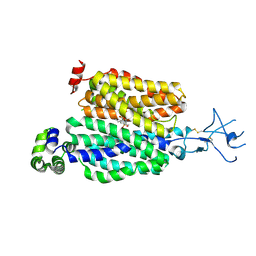 | |
8SDY
 
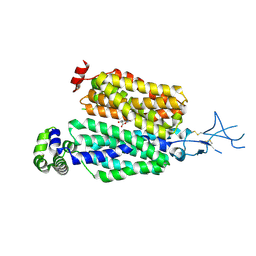 | |
6AYF
 
 | | TRPML3/ML-SA1 complex at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-3 | | Authors: | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2O52
 
 | | Crystal structure of human RAB4B in complex with GDP | | Descriptor: | BETA-MERCAPTOETHANOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhu, H, Tempel, W, Wang, J, Shen, Y, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB4B in complex with GDP
To be Published
|
|
4CDQ
 
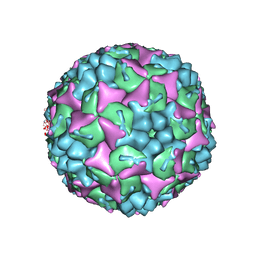 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4GZ5
 
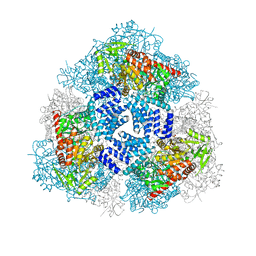 | | Crystal structure of human O-GlcNAc Transferase with UDP-GlcNAc | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
2OIL
 
 | | Crystal structure of human RAB25 in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-25, ... | | Authors: | Zhu, H, Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-11 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human RAB25 in complex with GDP
To be Published
|
|
4CEW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4GYW
 
 | | Crystal structure of human O-GlcNAc Transferase in complex with UDP and a glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
8AY2
 
 | |
5VVV
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, a-crystallin B | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
