6V2D
 
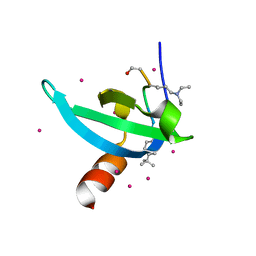 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V41
 
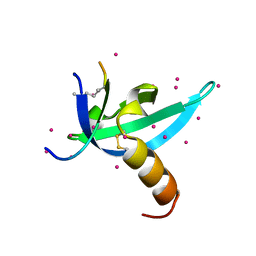 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
5HRW
 
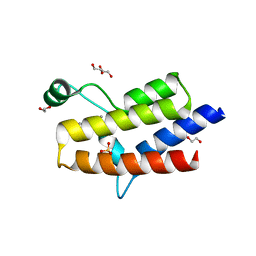 | | Crystal structure of the fifth bromodomain of human PB1 in complex with 1-propylisochromeno[3,4-c]pyrazol-5(2H)-one) compound | | Descriptor: | 1,2-ETHANEDIOL, 1-propylisochromeno[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1, ... | | Authors: | Tallant, C, Myrianthopoulos, V, Gaboriaud-Kolar, N, Newman, J.A, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Mikros, E, Knapp, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
5J0D
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with a benzoxazepine compound | | Descriptor: | 1,2-ETHANEDIOL, 7-(3,5-dimethoxyphenyl)-N-[(3S)-1-methylpiperidin-3-yl]-4-propanoyl-2,3,4,5-tetrahydro-1,4-benzoxazepine-9-carboxamide, CREB-binding protein | | Authors: | Tallant, C, Popp, T.A, Fedorov, O, Siejka, P, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-28 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Development of Selective CBP/P300 Benzoxazepine Bromodomain Inhibitors.
J.Med.Chem., 59, 2016
|
|
3QMH
 
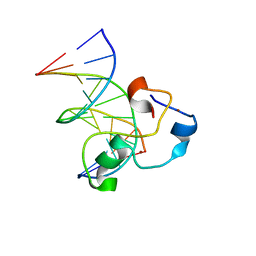 | | Structural Basis of Selective Binding of Non-Methylated CpG islands (DNA-TCGA) by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*CP*GP*AP*TP*GP*GP*C)-3', CpG-binding protein, ZINC ION | | Authors: | Xu, C, Bian, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
6W6D
 
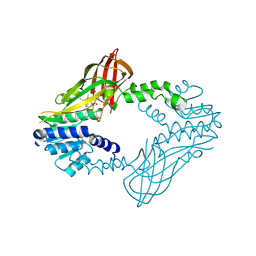 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
5E16
 
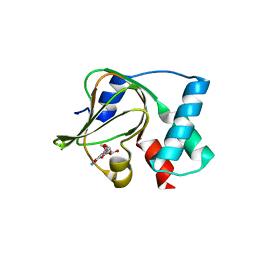 | | Co-crystal structure of the N-termial cGMP binding domain of Plasmodium falciparum PKG with cGMP | | Descriptor: | CGMP-dependent protein kinase, CYCLIC GUANOSINE MONOPHOSPHATE | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5VC9
 
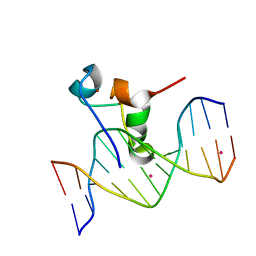 | | Zinc finger of human CXXC4 in complex with CpG DNA | | Descriptor: | CXXC-type zinc finger protein 4, CpG DNA, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
3N9V
 
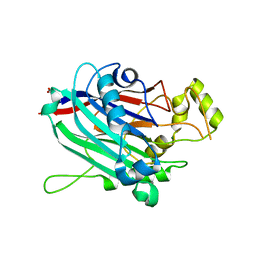 | | Crystal Structure of INPP5B | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
3QMG
 
 | | Structural Basis of Selective Binding of Non-Methylated CpG islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*AP*CP*GP*GP*TP*GP*GP*C)-3', 5'-D(*GP*CP*CP*AP*CP*CP*GP*TP*TP*GP*GP*C)-3', CpG-binding protein, ... | | Authors: | Xu, C, Bian, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3MTC
 
 | | Crystal Structure of INPP5B in complex with phosphatidylinositol 4-phosphate | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
6BHG
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHI
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHD
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHE
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
3QMI
 
 | | Structural Basis of Selective Binding of Non-Methylated CpG islands (DNA-ACGT) by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3', CpG-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xu, C, Bian, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
4CRI
 
 | | Crystal Structure of 53BP1 tandem tudor domains in complex with methylated K810 Rb peptide | | Descriptor: | RB1 PROTEIN, TUMOR SUPPRESSOR P53-BINDING PROTEIN 1 | | Authors: | Krojer, T, Johansson, C, Gileadi, C, Fedorov, O, Carr, S, La Thangue, N.B, Vollmar, M, Crawley, L, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-02-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lysine Methylation-Dependent Binding of 53BP1 to the Prb Tumor Suppressor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8P2W
 
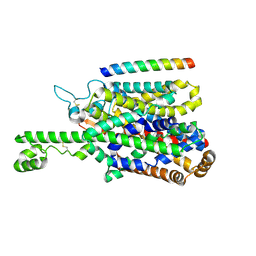 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Z
 
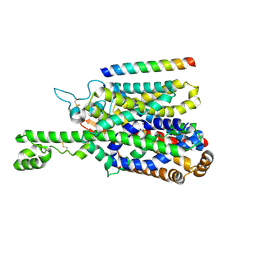 | | Structure of human SIT1 bound to L-pipecolate (focussed map / refinement) | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P30
 
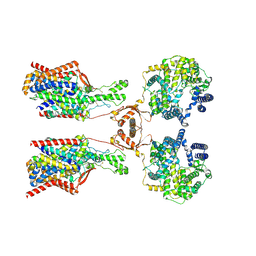 | | Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
3KVO
 
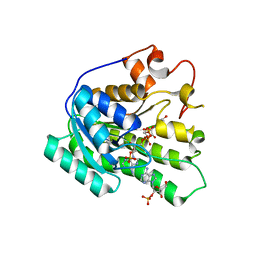 | | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2) | | Descriptor: | Hydroxysteroid dehydrogenase-like protein 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Bhatia, C, Huang, J, Pilka, E, Muniz, J.R.C, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Verdin, E.M, Oppermann, U, Kavanagh, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2)
To be Published
|
|
8P2Y
 
 | | Structure of human SIT1:ACE2 complex (closed PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P31
 
 | | Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2X
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
6ZH0
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
