6EWP
 
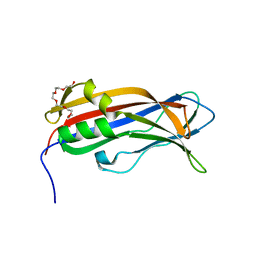 | | Mus musculus CEP120 third C2 domain (C2C) | | Descriptor: | Centrosomal protein of 120 kDa, HEXAETHYLENE GLYCOL | | Authors: | van Breugel, M, al-Jassar, C. | | Deposit date: | 2017-11-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Disease-Associated Mutations in CEP120 Destabilize the Protein and Impair Ciliogenesis.
Cell Rep, 23, 2018
|
|
6FAU
 
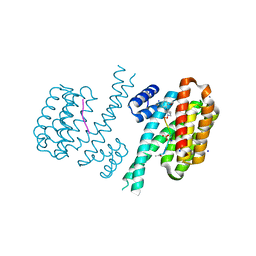 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2e-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(2-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6EZC
 
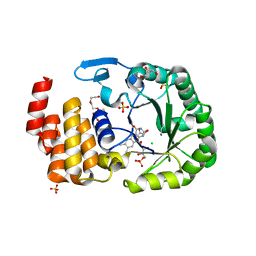 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain E294K Q305K double mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Bregeon, D, Vincent, G, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
6EZJ
 
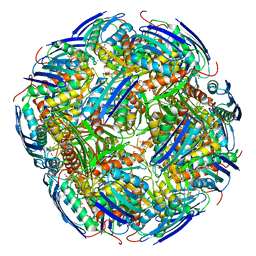 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FHV
 
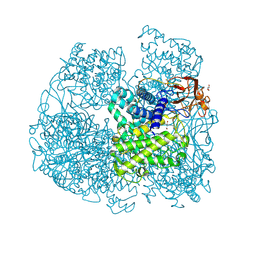 | | Crystal structure of Penicillium oxalicum Glucoamylase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FI4
 
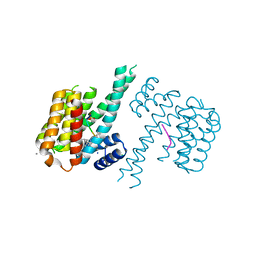 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2e with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FAV
 
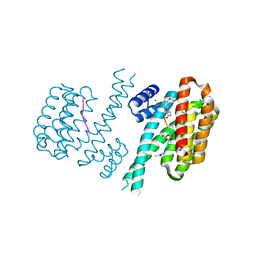 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FBY
 
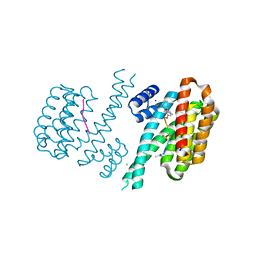 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2b with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methylphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6EWH
 
 | |
6EWL
 
 | |
8JIL
 
 | | Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
6FI5
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2d with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
6ZV6
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state B (post 18S rRNA cleavage) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-24 | | Release date: | 2021-05-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
6ZUO
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state A (pre 18S rRNA cleavage) | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
8JII
 
 | | Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JHY
 
 | | Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIM
 
 | | Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6PB1
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
1AFE
 
 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
6Y20
 
 | |
9D6E
 
 | | Gag CA-SP1 immature lattice bound with Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9D6D
 
 | |
9D88
 
 | |
