2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IQ1
 
 | | Crystal structure of human PPM1K | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C kappa, PPM1K | | Authors: | Bonanno, J.B, Freeman, J, Russell, M, Bain, K.T, Adams, J, Pelletier, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
6RIM
 
 | |
7YSX
 
 | | Crystal structure of PDE4D complexed with licoisoflavone A | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YQF
 
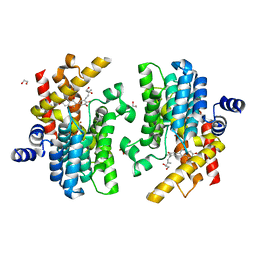 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
6PAV
 
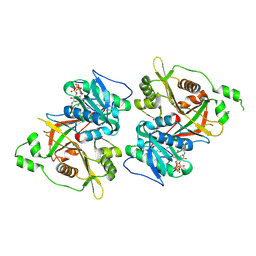 | |
7WKS
 
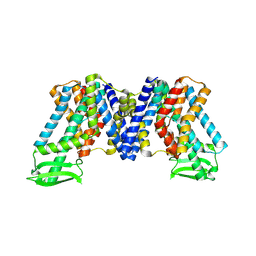 | | Apo state of AtPIN3 | | Descriptor: | Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
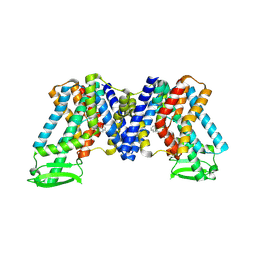 | | NPA bound state of AtPIN3 | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7MJU
 
 | |
6PAU
 
 | |
6JNX
 
 | | Cryo-EM structure of a Q-engaged arrested complex | | Descriptor: | Antiterminator Q protein, DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Feng, Y, Shi, J. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of Q-dependent transcription antitermination.
Nat Commun, 10, 2019
|
|
1RXD
 
 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | Descriptor: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | Authors: | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
8TBH
 
 | | Tricomplex of RMC-7977, KRAS G12R, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Bar Ziv, T, Zhang, D, Tomlinson, A.C.A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
7NAB
 
 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
6JNY
 
 | |
3R22
 
 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7XXB
 
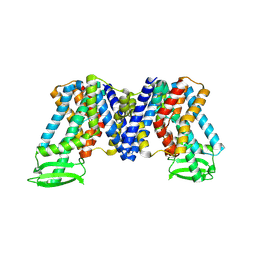 | | IAA bound state of AtPIN3 | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-05-29 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
2YIX
 
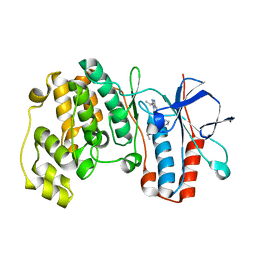 | | Triazolopyridine Inhibitors of p38 | | Descriptor: | 1-ethyl-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Han, s, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthawaite, R.A, Mahke, A, Marr, E, Mathias, J.P, Philip, J, Phillips, C, Smith, R.T, Stefaniak, M.H, Yeadon, M. | | Deposit date: | 2011-05-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
3R21
 
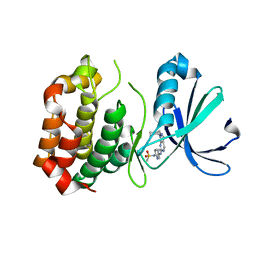 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
1GSU
 
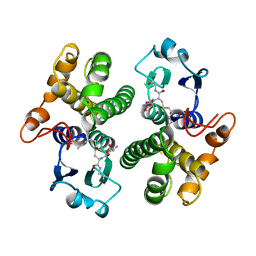 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | Descriptor: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | Deposit date: | 1997-09-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
