8VZK
 
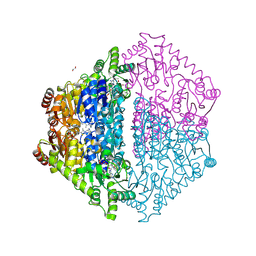 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase CfhHACS from Chloroflexi bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Gade, P, Mayfield, A, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
1CS7
 
 | | SYNTHETIC DNA HAIRPIN WITH STILBENEDIETHER LINKER | | Descriptor: | 5'-D(GP*(BRU)P*TP*TP*TP*GP*(S02)*CP*AP*AP*AP*AP*C)-3', STRONTIUM ION | | Authors: | Lewis, F.D, Liu, X, Wu, Y, Miller, S.E, Wasielewski, M.R, Letsinger, R.L, Sanishvili, R, Joachimiak, A, Tereshko, V, Egli, M. | | Deposit date: | 1999-08-17 | | Release date: | 2001-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Photoinduced Electron Transfer in Exceptionally Stable Synthetic DNA Hairpins with Stilbenediether Linkers
J.Am.Chem.Soc., 121, 1999
|
|
1D8X
 
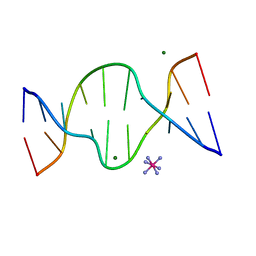 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*TP*GP*AP*GP*G)-3', COBALT HEXAMMINE(III), MAGNESIUM ION | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-10-26 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1DNO
 
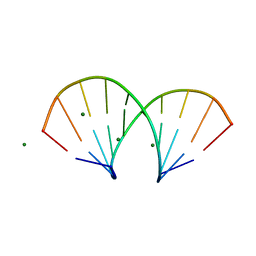 | | A-DNA/RNA DODECAMER R(GCG)D(TATACGC) MG BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP)-D(*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
1DCR
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1D9R
 
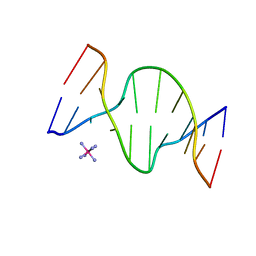 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', COBALT HEXAMMINE(III) | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-10-29 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1DNT
 
 | | RNA/DNA DODECAMER R(GC)D(GTATACGC) WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP*CP)-D(*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
1DNZ
 
 | | A-DNA DECAMER ACCGGCCGGT WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-17 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
1DNX
 
 | | RNA/DNA DODECAMER R(G)D(CGTATACGC) WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP)-D(*CP*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
1DPL
 
 | | A-DNA DECAMER GCGTA(T23)TACGC WITH INCORPORATED 2'-METHOXY-3'-METHYLENEPHOSPHATE-THYMIDINE | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(T23)P*AP*CP*GP*C)-3', MAGNESIUM ION, SPERMINE | | Authors: | Egli, M, Tereshko, V, Teplova, M, Minasov, G, Joachimiak, A, Sanishvili, R, Weeks, C.M, Miller, R, Maier, M.A, An, H, Dan Cook, P, Manoharan, M. | | Deposit date: | 1999-12-27 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution.
Biopolymers, 48, 1998
|
|
1DPN
 
 | | B-DODECAMER CGCGAA(TAF)TCGCG WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Teplova, M, Minasov, G, Joachimiak, A, Sanishvili, R, Weeks, C.M, Miller, R, Maier, M.A, An, H, Dan Cook, P, Manoharan, M. | | Deposit date: | 1999-12-27 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution.
Biopolymers, 48, 1998
|
|
6DFP
 
 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
1NG5
 
 | |
3LW7
 
 | | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylate kinase related protein (AdkA-like) | | Authors: | Stein, A.J, Sather, A, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A
To be Published
|
|
3LVT
 
 | | The Crystal Structure of a Protein in the Glycosyl Hydrolase Family 38 from Enterococcus faecalis to 2.55A | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 38 | | Authors: | Stein, A.J, Binkowski, T.A, Weger, A, Borovilos, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Crystal Structure of a Protein in the Glycosyl Hydrolase Family 38 from Enterococcus faecalis to 2.55A
To be Published
|
|
3LWA
 
 | | The Crystal Structure of a Secreted Thiol-disulfide Isomerase from Corynebacterium glutamicum to 1.75A | | Descriptor: | CALCIUM ION, Secreted thiol-disulfide isomerase | | Authors: | Stein, A.J, Weger, A, Hendricks, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of a Secreted Thiol-disulfide Isomerase from Corynebacterium glutamicum to 1.75A
To be Published
|
|
3M1A
 
 | | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A | | Descriptor: | ACETATE ION, Putative dehydrogenase, SODIUM ION | | Authors: | Stein, A.J, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A
To be Published
|
|
3MFN
 
 | | Dfer_2879 protein of unknown function from Dyadobacter fermentans | | Descriptor: | ACETATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Chin, S, Eisen, J, Wu, D, Kerfeld, C, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-02 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | X-ray crystal structure of Dfer_2879 protein of unknown function from Dyadobacter fermentans.
To be Published
|
|
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
6B6L
 
 | | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Glycosyl hydrolase family 2, ... | | Authors: | Tan, K, Joachimiak, G, Nocek, B, Enddres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
1P99
 
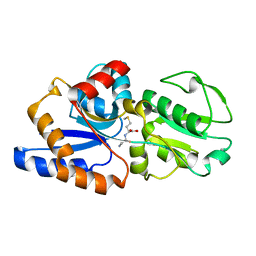 | | 1.7A crystal structure of protein PG110 from Staphylococcus aureus | | Descriptor: | GLYCINE, Hypothetical protein PG110, METHIONINE | | Authors: | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions.
Biochemistry, 43, 2004
|
|
1ILV
 
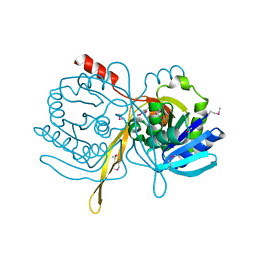 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
4MPT
 
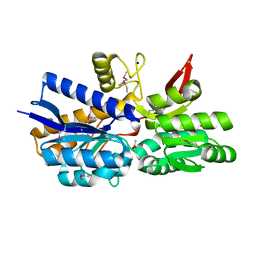 | | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I | | Descriptor: | ACETIC ACID, Putative leu/ile/val-binding protein, SODIUM ION | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I
To be Published
|
|
6VWW
 
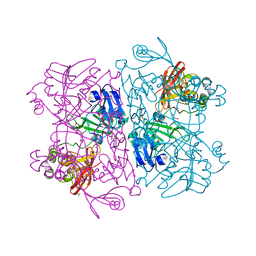 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
9EWK
 
 | | Solvent organization in ultrahigh-resolution protein crystal structure at room temperature | | Descriptor: | Crambin, ETHANOL | | Authors: | Chen, J.C.-H, Gilski, M, Chang, C, Borek, D, Rosenbaum, G, Lavens, A, Otwinowski, Z, Kubicki, M, Dauter, Z, Jaskolski, M, Joachimiak, A. | | Deposit date: | 2024-04-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Solvent organization in the ultrahigh-resolution crystal structure of crambin at room temperature.
Iucrj, 11, 2024
|
|
