7U0D
 
 | |
1Z1L
 
 | | The Crystal Structure of the Phosphodiesterase 2A Catalytic Domain | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Ding, Y.H, Kohls, D, Low, C. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants for Inhibitor Specificity and Selectivity in PDE2A Using the Wheat Germ in Vitro Translation System.
Biochemistry, 44, 2005
|
|
2MVT
 
 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | Descriptor: | Scoloptoxin SSD609 | | Authors: | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-09-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
1G72
 
 | | CATALYTIC MECHANISM OF QUINOPROTEIN METHANOL DEHYDROGENASE: A THEORETICAL AND X-RAY CRYSTALLOGRAPHIC INVESTIGATION | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE HEAVY SUBUNIT, METHANOL DEHYDROGENASE LIGHT SUBUNIT, ... | | Authors: | Zheng, Y, Xia, Z, Chen, Z, Bruice, T.C, Mathews, F.S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of quinoprotein methanol dehydrogenase: A theoretical and x-ray crystallographic investigation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5I5K
 
 | |
7WW2
 
 | | Structure of an Isocytosine specific deaminase Vcz | | Descriptor: | 8-oxoguanine deaminase, ZINC ION | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
4O0Y
 
 | | Back pocket flexibility provides group-II PAK selectivity for type 1 kinase inhibitors | | Descriptor: | 4-[1-(4-amino-1,3,5-triazin-2-yl)-2-(ethylamino)-1H-benzimidazol-6-yl]-2-methylbut-3-yn-2-ol, Serine/threonine-protein kinase PAK 4 | | Authors: | Rouge, L, Tam, C, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-02-12 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Back Pocket Flexibility Provides Group II p21-Activated Kinase (PAK) Selectivity for Type I 1/2 Kinase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4O0T
 
 | |
5J7J
 
 | |
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
3VNC
 
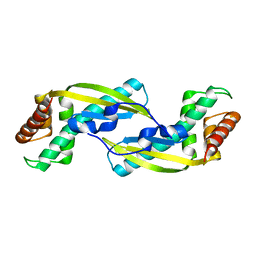 | | Crystal Structure of TIP-alpha N25 from Helicobacter Pylori in its natural dimeric form | | Descriptor: | TIP-alpha | | Authors: | Gao, M, Li, D, Hu, Y, Zou, Q, Wang, D.-C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of TNF-alpha-Inducing Protein from Helicobacter Pylori in Active Form Reveals the Intrinsic Molecular Flexibility for Unique DNA-Binding
Plos One, 7, 2012
|
|
5KCL
 
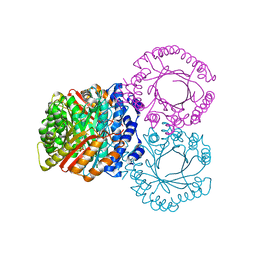 | |
5KCY
 
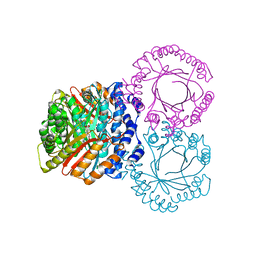 | |
4O0R
 
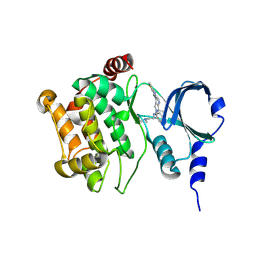 | |
5KD6
 
 | |
4O0V
 
 | |
5KCQ
 
 | |
5KDA
 
 | |
4PSQ
 
 | | Crystal Structure of Retinol-Binding Protein 4 (RBP4) in complex with a non-retinoid ligand | | Descriptor: | (1-benzyl-1H-imidazol-4-yl)[4-(2-chlorophenyl)piperazin-1-yl]methanone, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Wang, Z, Johnstone, S, Walker, N. | | Deposit date: | 2014-03-07 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-assisted discovery of the first non-retinoid ligands for Retinol-Binding Protein 4.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4O0X
 
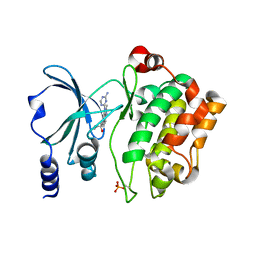 | | Back pocket flexibility provides group-II PAK selectivity for type 1 kinase inhibitors | | Descriptor: | 1-{[1-(4-amino-1,3,5-triazin-2-yl)-2-methyl-1H-benzimidazol-6-yl]ethynyl}cyclohexanol, Serine/threonine-protein kinase PAK 4 | | Authors: | Rouge, L, Tam, C, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Back Pocket Flexibility Provides Group II p21-Activated Kinase (PAK) Selectivity for Type I 1/2 Kinase Inhibitors.
J.Med.Chem., 57, 2014
|
|
5KCG
 
 | |
5KD0
 
 | |
