8FIH
 
 | |
8FIT
 
 | |
8FG6
 
 | |
7YDT
 
 | | Crystal structure of mouse MPND | | Descriptor: | MPN domain containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
7YDW
 
 | | Crystal structure of the MPND-DNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MPN domain-containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
8G1Q
 
 | | Co-crystal structure of Compound 1 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongin-B, Elongin-C, ... | | Authors: | Ghimire Rijal, S, Wurz, R.P, Vaish, A. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation.
Nat Commun, 14, 2023
|
|
8G1P
 
 | | Co-crystal structure of Compound 11 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Ghimire Rijal, S, Wurz, R.P, Vaish, A. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation.
Nat Commun, 14, 2023
|
|
8GA9
 
 | |
8FVT
 
 | |
8G8I
 
 | |
8GAQ
 
 | |
8GAA
 
 | |
8FWF
 
 | | Crystal structure of Apo form Fab235 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-21 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FXJ
 
 | | Crystal structure of Fab460 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab460, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FYM
 
 | | Crystal structure of Fab235 in complex with MPER peptide | | Descriptor: | ALA-SER-LEU-TRP-ASN-TRP-PHE-ASN-ILE-THR-ASN-TRP-LEU-TRP-TYR-ILE-LYS-LYS-LYS, CHLORIDE ION, Fab235, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FZ2
 
 | | Crystal structure of Fab460 in complex with MPER peptide | | Descriptor: | Fab460, H chain, L chain, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8GJG
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | gluc_A04_0005, gluc_A04_0005 Binder | | Authors: | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8H89
 
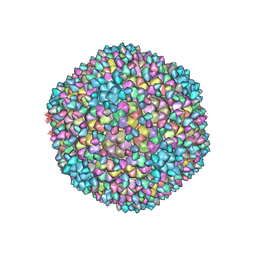 | | Capsid of Ralstonia phage GP4 | | Descriptor: | Major capsid protein, Virion associated protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2022-10-22 | | Release date: | 2022-11-16 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9.
Viruses, 14, 2022
|
|
8Y0W
 
 | | dormant ribosome with eIF5A, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
8Y0U
 
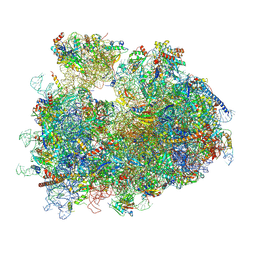 | | dormant ribosome with STM1 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
8FIQ
 
 | |
8FIN
 
 | |
8FUR
 
 | | Crystal structure of human IDO1 with compound 11 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-methylphenyl)-N'-[(1P,2'P)-4-propoxy-5-propyl-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-yl]urea | | Authors: | Critton, D.A, Lewis, H.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and biological evaluation of biaryl alkyl ethers as inhibitors of IDO1.
Bioorg.Med.Chem.Lett., 88, 2023
|
|
8GJI
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | GCG binder, Glucagon | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
