7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
3UA0
 
 | | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease | | Descriptor: | Fibroin heavy chain | | Authors: | He, Y.-X, Zhang, N.-N, Chen, B.-Y, Li, W.-F, Chen, Y.-X, Zhou, C.-Z. | | Deposit date: | 2011-10-20 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease.
J.Mol.Biol., 418, 2012
|
|
4HXZ
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HY1
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)-1H-isoindol-1-one, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2LUY
 
 | | Solution structure of the tandem zinc finger domain of fission yeast Stc1 | | Descriptor: | Meiotic chromosome segregation protein P8B7.28c, ZINC ION | | Authors: | He, C, Shi, Y, Bayne, E, Wu, J. | | Deposit date: | 2012-06-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
5ZMB
 
 | |
7W9N
 
 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | Authors: | Xu, G.H, Li, C.G. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
2MDJ
 
 | |
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
2MDC
 
 | | Solution structure of the WW domain of HYPB | | Descriptor: | Histone-lysine N-methyltransferase SETD2 | | Authors: | Gao, Y.G. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin
Structure, 22, 2014
|
|
4GNE
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
5XM8
 
 | |
8WI0
 
 | | wt-hMRP5 inward-open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI5
 
 | | M5PI-bound hMRP5 | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI3
 
 | | ND-hMRP5-inward open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI2
 
 | | RD-hMRP5-inward open | | Descriptor: | ATP-binding cassette sub-family C member 5, CYS-GLN-ASP-ALA-LEU-GLU-THR-ALA-ALA-ARG-ALA-GLU-GLY-LEU-SER-LEU-ASP-ALA-SER | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI4
 
 | | m6-hMRP5 inward open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
6L4K
 
 | | Human serum albumin-Palmitic acid-Cu compound | | Descriptor: | 2-bromanyl-9-ethyl-~{N},~{N},7-trimethyl-3-thia-1$l^{4},5,6$l^{4},10-tetraza-2$l^{4}-cupratricyclo[6.4.0.0^{2,6}]dodeca-1(8),4,6,9,11-pentaen-4-amine, PALMITIC ACID, Serum albumin | | Authors: | Zhang, Z.L. | | Deposit date: | 2019-10-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Brain-Tumor-Inhibiting Copper(II) Compound Based on a Human Serum Albumin (HSA)-Cell Penetrating Peptide Conjugate.
J.Med.Chem., 62, 2019
|
|
5XM9
 
 | |
4MQU
 
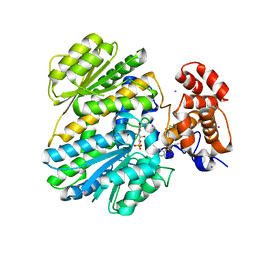 | | Human GKRP complexed to AMG-3969 and S6P | | Descriptor: | 2-{4-[(2S)-4-[(6-aminopyridin-3-yl)sulfonyl]-2-(prop-1-yn-1-yl)piperazin-1-yl]phenyl}-1,1,1,3,3,3-hexafluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | St Jean, D.J, Ashton, K.S, Bartberger, M.D, Chen, J, Chmait, S, Cupples, R, Galbreath, E, Helmering, J, Jordan, S.R, Liu, L. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 2. Leveraging structure-based drug design to identify analogues with improved pharmacokinetic profiles.
J.Med.Chem., 57, 2014
|
|
4LCH
 
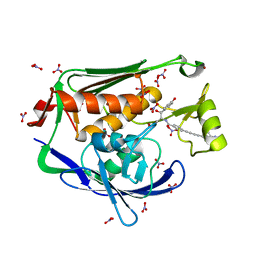 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | Descriptor: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
6LYC
 
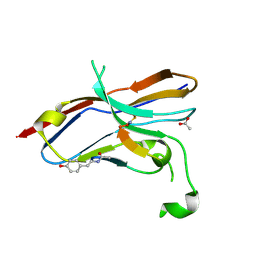 | | Crystal structure of the NOD SIRPa complex with D4-2 | | Descriptor: | ACETIC ACID, D4-2, SIRPa of the NOD mouse strain | | Authors: | Murata, Y, Matsuda, M, Nakagawa, A, Matozaki, T. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Cell Chem Biol, 27, 2020
|
|
