6YFO
 
 | | Virus-like particle of bacteriophage GQ-907 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFU
 
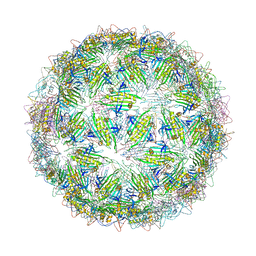 | | Virus-like particle of Wenzhou levi-like virus 4 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.018 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
4ACS
 
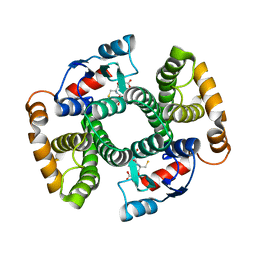 | | Crystal structure of mutant GST A2-2 with enhanced catalytic efficiency with azathioprine | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE A2 | | Authors: | Zhang, W, Moden, O, Tars, K, Mannervik, B. | | Deposit date: | 2011-12-19 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based redesign of GST A2-2 for enhanced catalytic efficiency with azathioprine.
Chem.Biol., 19, 2012
|
|
4ANG
 
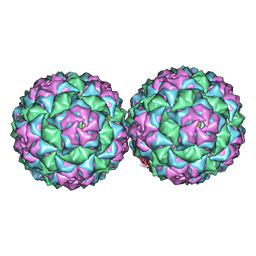 | | Small RNA phage PRR1 in complex with an RNA operator fragment | | Descriptor: | 5'-R(*CP*CP*AP*UP*AP*AP*GP*GP*AP*GP*CP*UP*AP*CP *CP*UP*AP*UP*GP*GP)-3', CALCIUM ION, COAT PROTEIN | | Authors: | Persson, M, Tars, K, Liljas, L. | | Deposit date: | 2012-03-16 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Prr1 Coat Protein Binding to its RNA Translational Operator
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7Q47
 
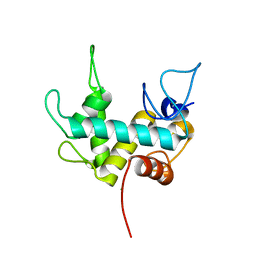 | | Endolysin from bacteriophage Enc34, catalytic domain | | Descriptor: | CHLORIDE ION, Endolysin, SODIUM ION | | Authors: | Cernooka, E, Rumnieks, J, Kazaks, A, Tars, K. | | Deposit date: | 2021-10-29 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diversity of the lysozyme fold: structure of the catalytic domain from an unusual endolysin encoded by phage Enc34.
Sci Rep, 12, 2022
|
|
7PP9
 
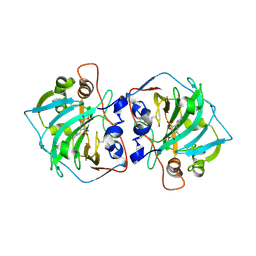 | |
2VF9
 
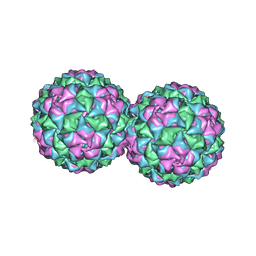 | |
2W4Z
 
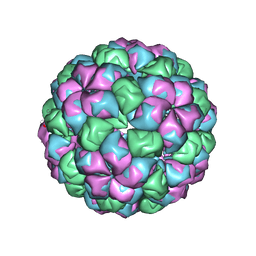 | | Caulobacter bacteriophage 5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, CAULOBACTER BACTERIOPHAGE 5 | | Authors: | Plevka, P, Kazaks, A, Dishlers, A, Liljas, L, Tars, K. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Structure of Bacteriophage Phicb5 Reveals a Role of the RNA Genome and Metal Ions in Particle Stability and Assembly.
J.Mol.Biol., 391, 2009
|
|
2YN7
 
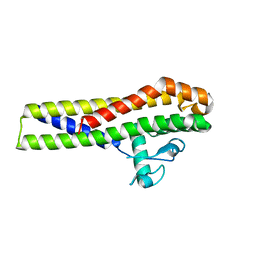 | | Crystal structure of an outer surface protein BBA66 from Borrelia burgdorferi | | Descriptor: | OUTER SURFACE PROTEIN | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Baumanis, V, Tars, K. | | Deposit date: | 2012-10-12 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Infectious Phenotype-Associated Outer Surface Protein Bba66 from the Lyme Disease Agent Borrelia Burgdorferi.
Ticks Tick Borne Dis., 5, 2014
|
|
2W4Y
 
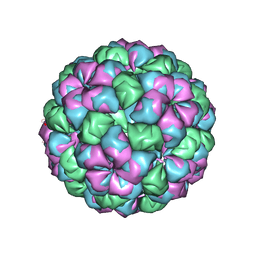 | | Caulobacter bacteriophage 5 - virus-like particle | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, CAULOBACTER 5 VIRUS-LIKE PARTICLE | | Authors: | Plevka, P, Kazaks, A, Dishlers, A, Liljas, L, Tars, K. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of Bacteriophage Phicb5 Reveals a Role of the RNA Genome and Metal Ions in Particle Stability and Assembly.
J.Mol.Biol., 391, 2009
|
|
2WBH
 
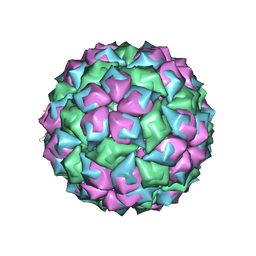 | |
2VTU
 
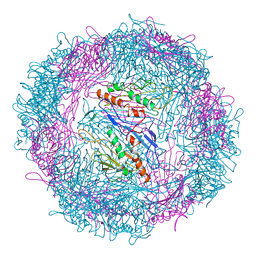 | |
8UW1
 
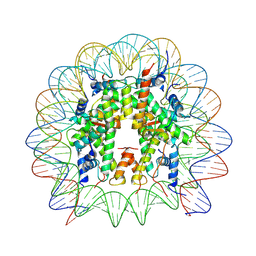 | | Cryo-EM structure of DNMT3A1 UDR in complex with H2AK119Ub-nucleosome | | Descriptor: | DNA (146-MER), DNA (cytosine-5)-methyltransferase 3A, Histone H2A, ... | | Authors: | Gretarsson, K, Abini-Agbomson, S, Armache, K.-J, Lu, C. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch.
Sci Adv, 10, 2024
|
|
5T87
 
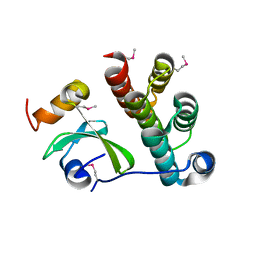 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
7A0O
 
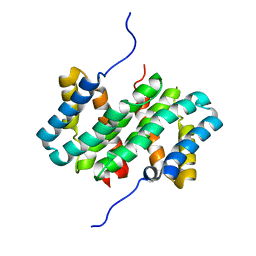 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 5.5 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7A0I
 
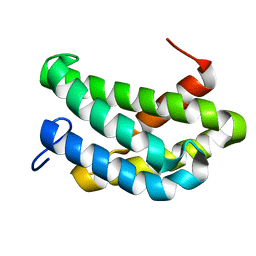 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 7.2 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7YZV
 
 | |
6QBI
 
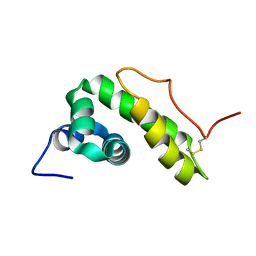 | | NMR structure of BB_P28, Borrelia burgdorferi outer surface lipoprotein | | Descriptor: | Surface protein, mlp lipoprotein family | | Authors: | Fridmanis, J, Otikovs, M, Brangulis, K, Jaudzems, K. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Borrelia burgdorferi outer surface lipoprotein BBP28, a member of the mlp protein family.
Proteins, 2020
|
|
3I6A
 
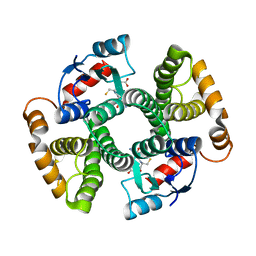 | | Human GST A1-1 GIMF mutant with Glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
3I69
 
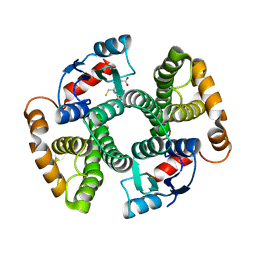 | | Apo Glutathione Transferase A1-1 GIMF-helix mutant | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
6QN1
 
 | | T=4 quasi-symmetric bacterial microcompartment particle | | Descriptor: | BMC domain-containing protein, Carbon dioxide concentrating mechanism protein CcmL | | Authors: | Kalnins, G. | | Deposit date: | 2019-02-08 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles.
Nat Commun, 11, 2020
|
|
6R6F
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-chloro-2-cyclohexylsulfanyl-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-27 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
6QS0
 
 | |
7PUW
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-chloro-4-[(2-phenylethyl)sulfanyl]-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7PUU
 
 | | Crystal structure of carbonic anhydrase XII with methyl 4-chloro-2-cyclohexylsulfanyl-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
