1C53
 
 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | Descriptor: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | Deposit date: | 1991-08-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
4JEA
 
 | | Crystal structure of an engineered Zn-RIDC1 construct with four interfacial disulfide bonds | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ZINC ION, ... | | Authors: | Tezcan, F.A, Medina-Morales, A.M, Perez, A, Brodin, J.D. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | In Vitro and Cellular Self-Assembly of a Zn-Binding Protein Cryptand via Templated Disulfide Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6EN1
 
 | |
2GXB
 
 | | Crystal Structure of The Za Domain bound to Z-RNA | | Descriptor: | 5'-R(P*(DU)P*CP*GP*CP*GP*CP*G)-3', Double-stranded RNA-specific adenosine deaminase, SODIUM ION | | Authors: | Athanasiadis, A, Placido, D, Rich, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Left-Handed RNA Double Helix Bound by the Zalpha Domain of the RNA-Editing Enzyme ADAR1.
Structure, 15, 2007
|
|
3TYR
 
 | | Crystal structure of transcriptional regulator VanUg, Form I | | Descriptor: | Transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Dong, A, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form I
TO BE PUBLISHED
|
|
6E65
 
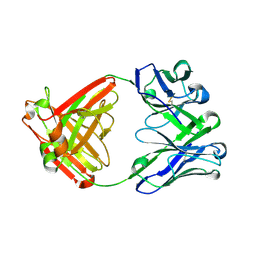 | |
4KNI
 
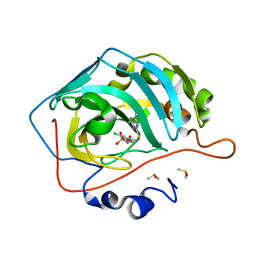 | | Crystal structure of human carbonic anhydrase isozyme II with 2-Chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2013-05-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII
Bioorg.Med.Chem., 21, 2013
|
|
4KP8
 
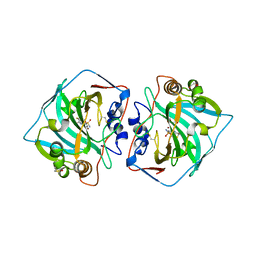 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with 3-[(Pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2013-05-13 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII
Bioorg.Med.Chem., 21, 2013
|
|
2HU6
 
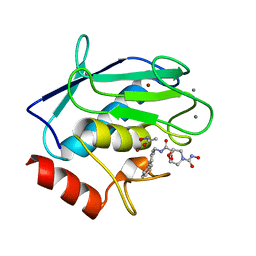 | | Crystal structure of human MMP-12 in complex with acetohydroxamic acid and a bicyclic inhibitor | | Descriptor: | (1S,5S,7R)-N~7~-(BIPHENYL-4-YLMETHYL)-N~3~-HYDROXY-6,8-DIOXA-3-AZABICYCLO[3.2.1]OCTANE-3,7-DICARBOXAMIDE, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Mannino, C, Nievo, M, Machetti, F, Papakyriakou, A, Calderone, V, Fragai, M, Guarna, A. | | Deposit date: | 2006-07-26 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Synthesis of bicyclic molecular scaffolds (BTAa): an investigation towards new selective MMP-12 inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
3TUS
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXYBENZOIC ACID, ... | | Authors: | Shukla, P.K, Gautam, L, Singh, A, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution
To be Published
|
|
2BAP
 
 | |
5DR1
 
 | |
2B0O
 
 | | Crystal structure of UPLC1 GAP domain | | Descriptor: | UPLC1, ZINC ION | | Authors: | Ismail, S, Shen, L, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural analysis of GAP and ankyrin domains of UPLC1
To be Published
|
|
3BWJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor lead compound Arc-1034 | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-N-(6-{[(1R)-4-carbamimidamido-1-{[(1R)-4-carbamimidamido-1-carbamoylbutyl]carbamoyl}butyl]amino}-6-oxohexyl)-3,4-dihydroxytetrahydrofuran-2-carboxamide, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Lavogina, D, Koenig, N, Uri, A, Bossemeyer, D. | | Deposit date: | 2008-01-09 | | Release date: | 2009-02-03 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases.
J.Med.Chem., 52, 2009
|
|
6H0M
 
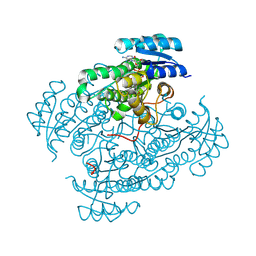 | | 17beta-hydroxysteroid dehydrogenase type 14 mutant K158A in complex with Nicotinamide Adenine Dinucleotide | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, DIMETHYL SULFOXIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Badran, M, Klebe, G, Heine, A, Marchais-Oberwinkler, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 17beta-hydroxysteroid dehydrogenase type 14 mutant K158A in complex with Nicotinamide Adenine Dinucleotide
To Be Published
|
|
6H0P
 
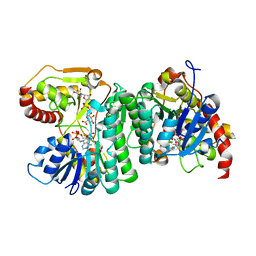 | |
2BBJ
 
 | | Crystal structure of the CorA Mg2+ transporter | | Descriptor: | divalent cation transport-related protein | | Authors: | Lunin, V.V, Dobrovetsky, E, Khutoreskaya, G, Zhang, R, Joachimiak, A, Bochkarev, A, Maguire, M.E, Edwards, A.M, Koth, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the CorA Mg2+ transporter
Nature, 440, 2006
|
|
6GOM
 
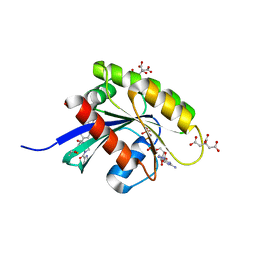 | | KRAS-169 Q61H GPPNHP + PPIN-1 | | Descriptor: | (6~{S})-1-(1~{H}-imidazol-4-ylcarbonyl)-6-[(4-phenylphenyl)methyl]-4-propyl-1,4-diazepan-5-one, CITRIC ACID, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3DYH
 
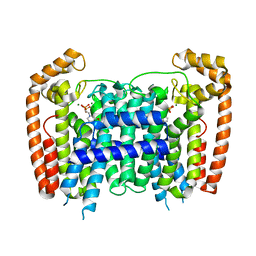 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-721 | | Descriptor: | 3-butoxy-1-(2,2-diphosphonoethyl)pyridinium, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
1Q71
 
 | | The structure of microcin J25 is a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Clark, R, Daly, N.L, Goransson, U, Jones, A, Craik, D.J. | | Deposit date: | 2003-08-14 | | Release date: | 2003-12-16 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone.
J.Am.Chem.Soc., 125, 2003
|
|
2B99
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase Complex with a Substrate analog inhibitor | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
5DR0
 
 | | Endothiapepsin in complex with fragment 203 | | Descriptor: | 1,2-ETHANEDIOL, 5-(methylsulfanyl)-4-(propan-2-ylsulfonyl)-1H-pyrazol-3-amine, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
5EAO
 
 | |
3RM0
 
 | | Human Thrombin in complex with MI354 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
