6VEE
 
 | |
6VED
 
 | | Solution structure of the TTD and linker region of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1 | | Authors: | Lemak, A, Houliston, S, Duan, S, Ong, M.S, Arrowsmith, C.H. | | Deposit date: | 2019-12-31 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
5MW5
 
 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protein jagged-2 | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
6VM6
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 2'3'3'-cAAA | | Descriptor: | 2'-5'-Linked Cyclic RNA (5'-R(P*AP*AP*A)-3'), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
5NOB
 
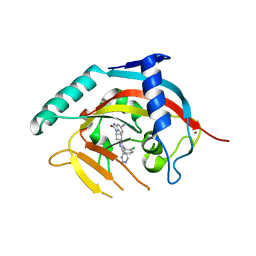 | | Crystal structure of human tankyrase 2 in complex with OD336 | | Descriptor: | 1-[3-[4-(2-chlorophenyl)-5-pyrimidin-4-yl-1,2,4-triazol-3-yl]cyclobutyl]-2-oxidanylidene-3~{H}-benzimidazole-5-carbonitrile, Tankyrase-2, ZINC ION | | Authors: | Ignatev, A, Lehtio, L. | | Deposit date: | 2017-04-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Novel Series of Tankyrase Inhibitors by a Hybridization Approach.
J. Med. Chem., 60, 2017
|
|
5N0J
 
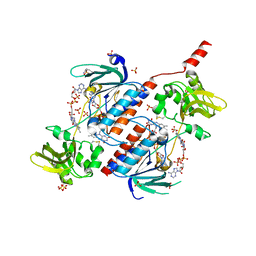 | | Structure of a novel oxidoreductase from Gloeobacter violaceus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Gll2934 protein, SULFATE ION | | Authors: | Buey, R.M, Galindo-Trigo, S, de Pereda, J.M, Balsera, M. | | Deposit date: | 2017-02-03 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Unprecedented pathway of reducing equivalents in a diflavin-linked disulfide oxidoreductase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MJY
 
 | |
6VM5
 
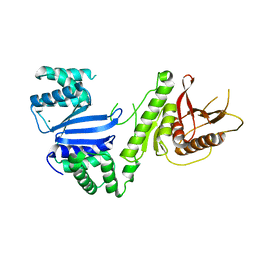 | | Structure of Moraxella osloensis Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | MAGNESIUM ION, SAVED domain-containing protein | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
8DCN
 
 | |
8DCM
 
 | |
5M4A
 
 | | Neutral trehalase Nth1 from Saccharomyces cerevisiae in complex with trehalose | | Descriptor: | Neutral trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Smidova, A, Alblova, M, Obsilova, V, Obsil, T. | | Deposit date: | 2016-10-18 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WAM
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
5M3V
 
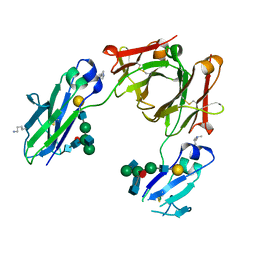 | | BEAT Fc | | Descriptor: | Ig gamma-1 chain C region, Ig gamma-1 chain C region,Ig gamma-3 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Skegro, D, Stutz, C, Bourquin, F, Blein, S. | | Deposit date: | 2016-10-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Immunoglobulin domain interface exchange as a platform technology for the generation of Fc heterodimers and bispecific antibodies.
J. Biol. Chem., 292, 2017
|
|
5NIS
 
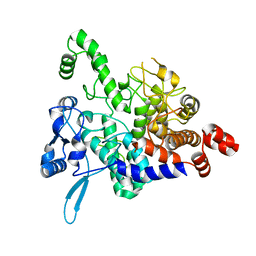 | |
5N6N
 
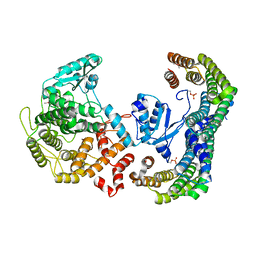 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | Descriptor: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | Authors: | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | Deposit date: | 2017-02-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WAN
 
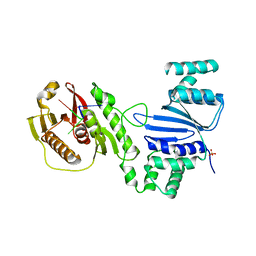 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6X2I
 
 | |
6WFU
 
 | |
5G1D
 
 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNDECAN-4, SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
5G1E
 
 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
7E48
 
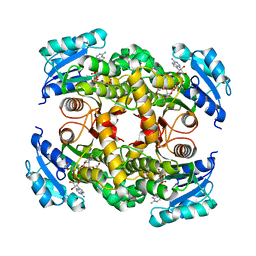 | | Crystal structure of InhA in complex with 3-nitropropanoic acid inhibitor | | Descriptor: | 3-NITROPROPANOIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Songsiriritthigul, C, Hanwarinroj, C, Suttipanta, N, Kamsri, P, Kittakoop, P, Pungpo, P. | | Deposit date: | 2021-02-10 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis InhA by 3-nitropropanoic acid.
Proteins, 90, 2022
|
|
8AVU
 
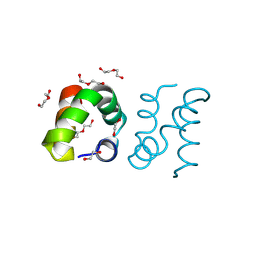 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the dimeric state | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin aureocin A53, D-Aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
8AVS
 
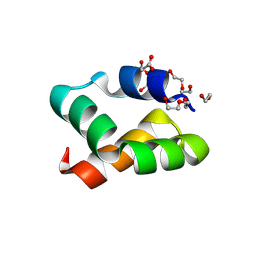 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the presence of citrate and acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bacteriocin aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
8AVT
 
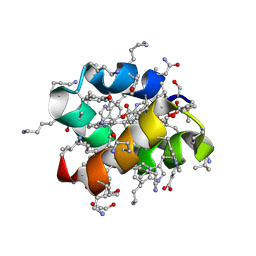 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the presence of glycerol 3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin aureocin A53, D-Aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
8AVR
 
 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the presence of sulfate | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin aureocin A53, D-Aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
