2EG4
 
 | | Crystal Structure of Probable Thiosulfate Sulfurtransferase | | Descriptor: | Probable thiosulfate sulfurtransferase, SULFATE ION, ZINC ION | | Authors: | Sakai, H, Ebihara, A, Kitamura, Y, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Probable Thiosulfate Sulfurtransferase
To be Published
|
|
3ASX
 
 | | Human Squalene synthase in complex with 1-{4-[{4-chloro-2-[(2-chlorophenyl)(hydroxy)methyl]phenyl}(2,2-dimethylpropyl)amino]-4-oxobutanoyl}piperidine-3-carboxylic acid | | Descriptor: | (3R)-1-{4-[{4-chloro-2-[(S)-(2-chlorophenyl)(hydroxy)methyl]phenyl}(2,2-dimethylpropyl)amino]-4-oxobutanoyl}piperidine-3-carboxylic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Shimizu, H, Suzuki, M, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
6QJB
 
 | | Truncated Evasin-3 (tEv3 17-56) | | Descriptor: | Evasin-3 | | Authors: | Denisov, S.S, Ippel, J.H, Heinzman, A.C.A, Koenen, R.R, Ortega-Gomez, A, Soehnlein, O, Hackeng, T.M, Dijkgraaf, I. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tick saliva protein Evasin-3 modulates chemotaxis by disrupting CXCL8 interactions with glycosaminoglycans and CXCR2.
J.Biol.Chem., 294, 2019
|
|
6GED
 
 | | Adhesin domain of PrgB from Enterococcus faecalis bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*GP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*CP*GP*GP*CP*CP*CP*G)-3'), ... | | Authors: | Schmitt, A, Berntsson, R.P.A. | | Deposit date: | 2018-04-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction.
Mol. Microbiol., 109, 2018
|
|
6QEX
 
 | |
3BUH
 
 | | BACE-1 complexed with compound 4 | | Descriptor: | 4-(2-aminoethyl)-2-cyclohexylphenol, beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6FNE
 
 | | Structure of human Brag2 (Sec7-PH domains) with the inhibitor Bragsin bound to the PH domain | | Descriptor: | (2~{S})-6-methyl-5-nitro-2-(trifluoromethyl)-2,3-dihydrochromen-4-one, IQ motif and SEC7 domain-containing protein 1, NONAETHYLENE GLYCOL | | Authors: | Nawrotek, A, Zeghouf, M, Cherfils, J. | | Deposit date: | 2018-02-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | PH-domain-binding inhibitors of nucleotide exchange factor BRAG2 disrupt Arf GTPase signaling.
Nat.Chem.Biol., 15, 2019
|
|
6QEP
 
 | | EngBF DARPin Fusion 4b H14 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MANGANESE (II) ION, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-01-08 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
3C8X
 
 | | Crystal structure of the ligand binding domain of human Ephrin A2 (Epha2) receptor protein kinase | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6QFO
 
 | | EngBF DARPin Fusion 9b 3G124 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MANGANESE (II) ION, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
2V1V
 
 | |
6QKY
 
 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2I4T
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with Imm-A | | Descriptor: | 3,4-PYRROLIDINEDIOL,2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)-2S,3S,4R,5R, PHOSPHATE ION, Trichomonas vaginalis purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Schramm, V.L, Almo, S.C. | | Deposit date: | 2006-08-22 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues.
Biochemistry, 46, 2007
|
|
2I81
 
 | | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced | | Descriptor: | 2-Cys Peroxiredoxin | | Authors: | Artz, J.D, Qiu, W, Dong, A, Lew, J, Ren, H, Zhao, Y, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced
To be published
|
|
3SHA
 
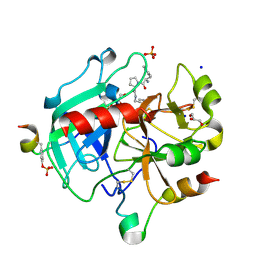 | | Human Thrombin In Complex With UBTHR97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(4-chloropyridin-3-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-06-16 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
6FXR
 
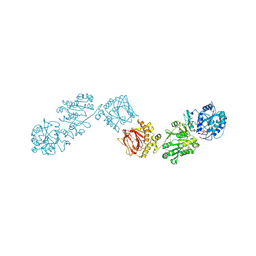 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Gal | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
3C3D
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and phosphate. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, PHOSPHATE ION | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
1JAC
 
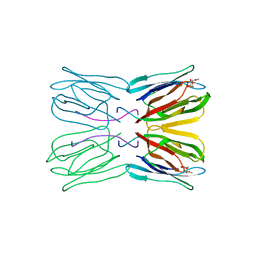 | | A NOVEL MODE OF CARBOHYDRATE RECOGNITION IN JACALIN, A MORACEAE PLANT LECTIN WITH A BETA-PRISM | | Descriptor: | JACALIN, methyl alpha-D-galactopyranoside | | Authors: | Sankaranarayanan, R, Sekar, K, Banerjee, R, Sharma, V, Surolia, A, Vijayan, M. | | Deposit date: | 1996-05-22 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold.
Nat.Struct.Biol., 3, 1996
|
|
6QJF
 
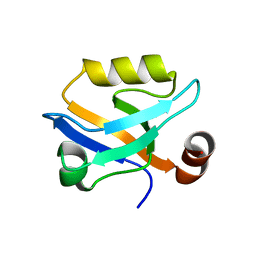 | |
6QJL
 
 | |
2HFV
 
 | | Solution NMR Structure of Protein RPA1041 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT90. | | Descriptor: | Hypothetical Protein RPA1041 | | Authors: | Eletsky, A, Atreya, H.S, Liu, G, Sukumaran, D, Garcia, M, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Pseudomonas aeruginosa Hypothetical Protein RPA1041
TO BE PUBLISHED
|
|
5EHI
 
 | |
6QL0
 
 | |
6QJG
 
 | |
6QJK
 
 | |
