6M8B
 
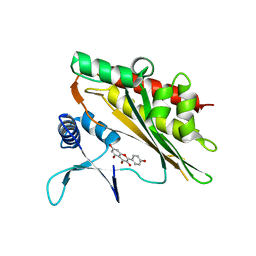 | |
8H3T
 
 | | The crystal structure of AlpH | | Descriptor: | AlpH, GLYCEROL | | Authors: | Zhao, Y, Li, M, Jiang, M, Pan, L.F. | | Deposit date: | 2022-10-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis.
Nat Commun, 14, 2023
|
|
6M1V
 
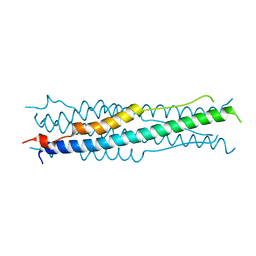 | |
6M8C
 
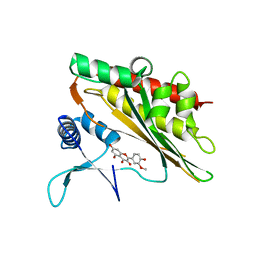 | |
6M8E
 
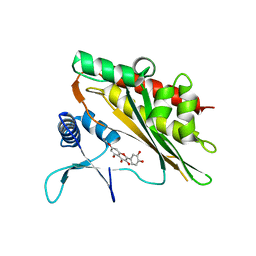 | |
6M8A
 
 | |
3SHQ
 
 | | Crystal Structure of UBLCP1 | | Descriptor: | MAGNESIUM ION, UBLCP1 | | Authors: | Xiao, J, Engel, J.L. | | Deposit date: | 2011-06-16 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MK8
 
 | |
6M88
 
 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2018-08-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of Inositol Polyphosphate Kinases by Quercetin and Related Flavonoids: A Structure-Activity Analysis.
J. Med. Chem., 62, 2019
|
|
2G46
 
 | |
7WGB
 
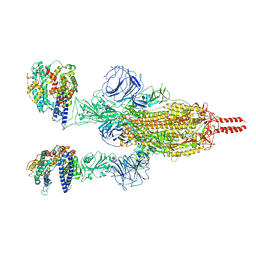 | | Neutral Omicron Spike Trimer in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG8
 
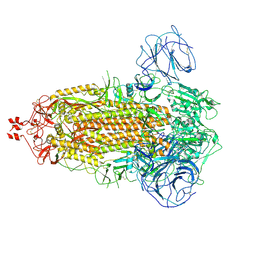 | | Delta Spike Trimer(3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
5ZMN
 
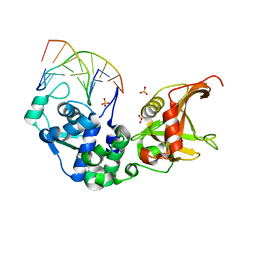 | | Sulfur binding domain and SRA domain of ScoMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*(GS)P*CP*CP*GP*GP*G)-3'), SULFATE ION, Uncharacterized protein McrA | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
5ZMM
 
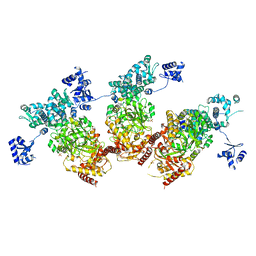 | | Structure of the Type IV phosphorothioation-dependent restriction endonuclease ScoMcrA | | Descriptor: | SULFATE ION, Uncharacterized protein McrA, ZINC ION | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
4OU6
 
 | | Crystal structure of DnaT84-153-dT10 ssDNA complex form 1 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | Authors: | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
7JJJ
 
 | | Structure of SARS-CoV-2 3Q-2P full-length dimers of spike trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
4OU7
 
 | | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | Authors: | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
4OUS
 
 | | Crystal structure of zebrafish Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2 | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
6A7A
 
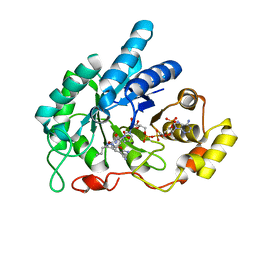 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
7XXY
 
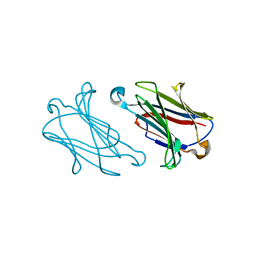 | | Macaca mulatta galectin-10/Charcot-Leyden crystal protein with lactose | | Descriptor: | Galectin-10/Charcot-Leyden crystal protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7XXW
 
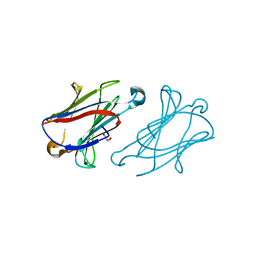 | |
7XXV
 
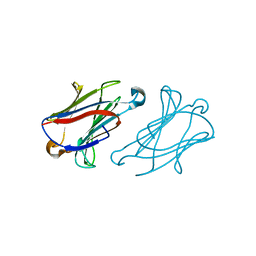 | | Macaca fascicularis galectin-10/Charcot-Leyden crystal protein with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7XXU
 
 | | Macaca fascicularis galectin-10/Charcot-Leyden crystal protein | | Descriptor: | Galectin | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7XXX
 
 | | Macaca mulatta galectin-10/Charcot-Leyden crystal protein | | Descriptor: | Galectin-10/Charcot-Leyden crystal protein | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7XXZ
 
 | | Macaca mulatta galectin-10/Charcot-Leyden crystal protein with glycerol | | Descriptor: | GLYCEROL, Galectin-10/Charcot-Leyden crystal protein | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
