4RZG
 
 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
4JIH
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Epalrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-[(2E)-2-methyl-3-phenylprop-2-en-1-ylidene]-4-oxo-2-thioxo-1,3-thiazolidin-3-yl}acetic acid | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JIR
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Epalrestat | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X. | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
8H01
 
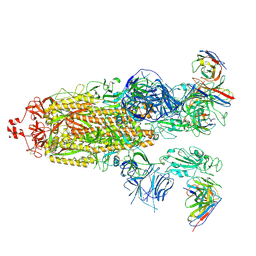 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8BYX
 
 | |
4Y73
 
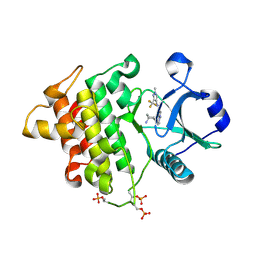 | | Crystal structure of IRAK4 kinase domain with inhibitor | | Descriptor: | 5-{[(1R,2S)-2-aminocyclohexyl]amino}-N-[1-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Lesburg, C.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of 5-Amino-N-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide Inhibitors of IRAK4.
Acs Med.Chem.Lett., 6, 2015
|
|
8H00
 
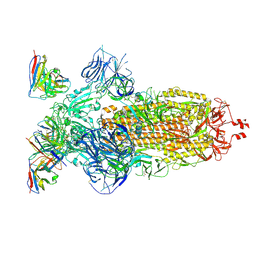 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8HEC
 
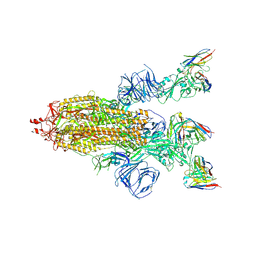 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8GZZ
 
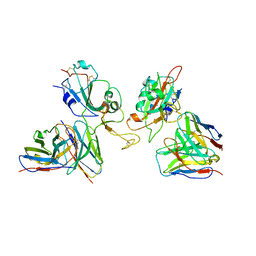 | | Local refinement of SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab | | Descriptor: | Spike protein S1, rabbit monoclonal antibody 1H1 Fab heavy chain, rabbit monoclonal antibody 1H1 Fab light chain | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEB
 
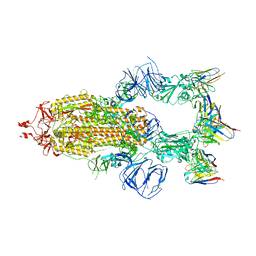 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
4WV5
 
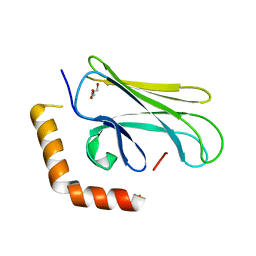 | |
4WV7
 
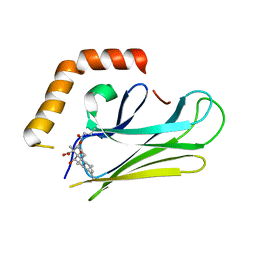 | | HEAT SHOCK PROTEIN 70 SUBSTRATE BINDING DOMAIN WITH COVALENTLY LINKED NOVOLACTONE | | Descriptor: | (5beta,6alpha,8alpha,14alpha)-13-ethenyl-5,6-dihydroxy-14-methylpodocarp-12-en-15-oic acid, Heat shock 70 kDa protein 1A/1B | | Authors: | Kirby, C.A, Baird, J, Stams, T. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The novolactone natural product disrupts the allosteric regulation of hsp70.
Chem.Biol., 22, 2015
|
|
4JII
 
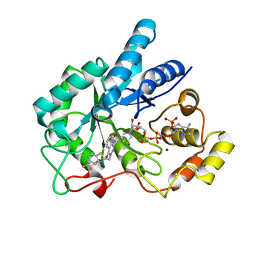 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
5GPK
 
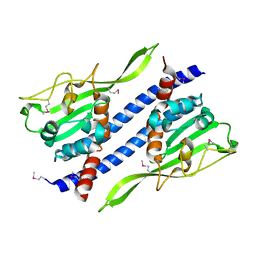 | | Crystal structure of Ccp1 mutant | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
3DPC
 
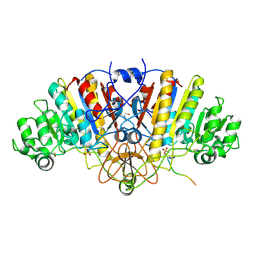 | |
9LO7
 
 | | The crystal structure of PDE4D with 2317b | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huag, Y.-Y, Wu, D, Luo, H.-B. | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.20006251 Å) | | Cite: | Structure-Based Optimization of Moracin M as Potent and Selective PDE4 Inhibitors with Antipsoriasis Effects.
J.Med.Chem., 68, 2025
|
|
4MQ2
 
 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
9IRY
 
 | | Structure of human URAT1 bound with verinurad | | Descriptor: | Solute carrier family 22 member 12, verinurad | | Authors: | Guo, W.J, Wei, M, Chen, L. | | Deposit date: | 2024-07-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of urate transport and uricosuric drugs inhibition in human URAT1.
Nat Commun, 16, 2025
|
|
9IRW
 
 | | Structure of human URAT1 bound with urate | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Guo, W.J, Wei, M, Chen, L. | | Deposit date: | 2024-07-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Mechanisms of urate transport and uricosuric drugs inhibition in human URAT1.
Nat Commun, 16, 2025
|
|
9IRX
 
 | | Structure of human URAT1 bound with benzbromarone | | Descriptor: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Guo, W.J, Wei, M, Chen, L. | | Deposit date: | 2024-07-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanisms of urate transport and uricosuric drugs inhibition in human URAT1.
Nat Commun, 16, 2025
|
|
4MQ1
 
 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, N-(5-{[(1R)-3-amino-1-(3-chlorophenyl)propyl]carbamoyl}-2-chlorophenyl)-2-methoxy-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, PENTAETHYLENE GLYCOL, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8GU4
 
 | | Poly(ethylene terephthalate) hydrolase (IsPETase)-linker | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
8GU5
 
 | | Wild type poly(ethylene terephthalate) hydrolase | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
