8HVH
 
 | | Cryo-EM structure of ABC transporter ABCC3 | | Descriptor: | ATP-binding cassette sub-family C member 3 | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
8HW2
 
 | | Cryo-EM structure of beta-estradiol 17-(beta-D-glucuronide)-bound human ABC transporter ABCC3 in nanodiscs | | Descriptor: | ATP-binding cassette sub-family C member 3, Estradiol-17beta-glucuronide | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
5CD3
 
 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
5CD5
 
 | |
8I91
 
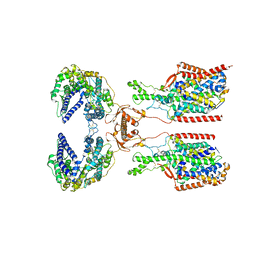 | | ACE2-SIT1 complex bound with proline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
9LTK
 
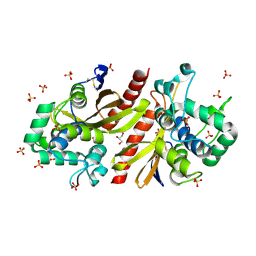 | | Crystal structur of Lpg1618(R215F) from Legionella pneumophila | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Liu, T, Gao, J, Ge, H. | | Deposit date: | 2025-02-06 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular characterization of AmpS, a class D beta-lactamase from Legionella pneumophila.
Int.J.Biol.Macromol., 312, 2025
|
|
9LSM
 
 | | The crystal structure of PDE5A with L9 | | Descriptor: | 1-[(13~{S},15~{R})-4-bromanyl-15-(3-chloranyl-4-methoxy-phenyl)-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaen-12-yl]ethanone, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
8I93
 
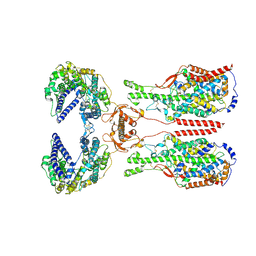 | | ACE2-B0AT1 complex bound with methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
8I92
 
 | | ACE2-B0AT1 complex bound with glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
9LSL
 
 | | The crystal structure of PDE5A with L1 | | Descriptor: | (13~{S},15~{R})-15-(3-chloranyl-4-methoxy-phenyl)-12-ethanoyl-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaene-4-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
8HTD
 
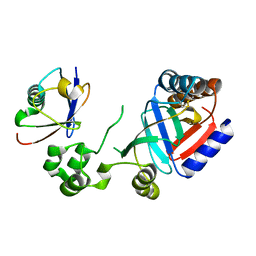 | | Crystal structure of an effector from Chromobacterium violaceum in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTE
 
 | | Crystal structure of an effector mutant in complex with ubiquitin | | Descriptor: | GLYCEROL, NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE, ... | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTF
 
 | | Crystal structure of an effector in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTC
 
 | | Crystal structure of a SeMet-labeled effector from Chromobacterium violaceum in complex with Ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
6KZK
 
 | |
2M75
 
 | |
2M7H
 
 | |
2M7F
 
 | |
8D1W
 
 | | Crystal structure of Plasmodium falciparum GRP78 in complex with (2R,3R,4S,5R)-2-(6-amino-8-((2-chlorobenzyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 8-{[(2-chlorophenyl)methyl]amino}adenosine, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W, Smil, D, Zepeda, C.A.V. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Plasmodium falciparum GRP78: nucleoside analogues as agents against the malaria chaperone
Frontiers in Molecular Bioscience, 2022
|
|
4RZF
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
8Z0F
 
 | | Cryo-EM structure of ATP-bound human very long-chain fatty acid ABC transporter ABCD3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 3 | | Authors: | Li, Y, Chen, Y.X, Zhou, C.Z, Hou, W.T. | | Deposit date: | 2024-04-09 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into human ABCD3-mediated peroxisomal acyl-CoA translocation.
Cell Discov, 10, 2024
|
|
9J66
 
 | |
8Z9X
 
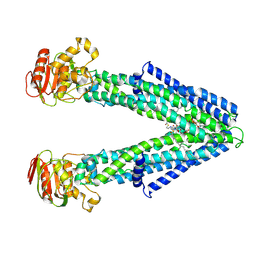 | | Cryo-EM structure of Phytanoyl-CoA-bound human very long-chain fatty acid ABC transporter ABCD3 | | Descriptor: | ATP-binding cassette sub-family D member 3, phytanoyl-CoA | | Authors: | Li, Y, Chen, Y.X, Zhou, C.Z, Hou, W.T. | | Deposit date: | 2024-04-23 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into human ABCD3-mediated peroxisomal acyl-CoA translocation.
Cell Discov, 10, 2024
|
|
5GO3
 
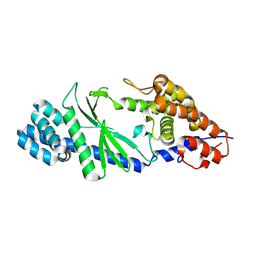 | | Crystal structure of a di-nucleotide cyclase Vibrio mutant | | Descriptor: | Cyclic GMP-AMP synthase | | Authors: | Ming, Z.H, Wang, W, Xie, Y.C, Chen, Y.C, Yan, L.M, Lou, Z.Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a di-nucleotide cyclase Vibrio mutant
To Be Published
|
|
5H3R
 
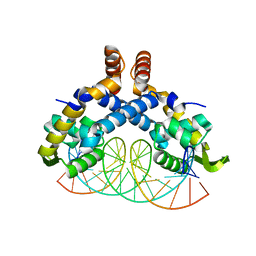 | | Crystal Structure of mutant MarR C80S from E.coli complexed with operator DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*CP*TP*TP*GP*CP*CP*TP*GP*GP*GP*CP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*TP*AP*TP*TP*GP*CP*CP*CP*AP*GP*GP*CP*AP*AP*GP*TP*AP*T)-3'), Multiple antibiotic resistance protein MarR | | Authors: | Zhu, R, Lou, H, Hao, Z. | | Deposit date: | 2016-10-26 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of the DNA-binding mechanism underlying the copper(II)-sensing MarR transcriptional regulator.
J. Biol. Inorg. Chem., 22, 2017
|
|
