8TQZ
 
 | | Eukaryotic translation initiation factor 2B with a mutation (L516A) in the delta subunit | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Lawrence, R, Sangwan, S, Anand, A, Shoemaker, S, Deal, A, Marqusee, S, Watler, P. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nat.Chem.Biol., 20, 2024
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
7TRJ
 
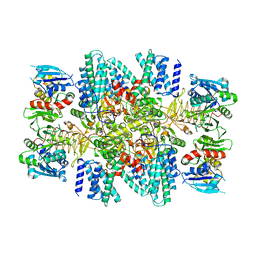 | | The eukaryotic translation initiation factor 2B from Homo sapiens with a H160D mutation in the beta subunit | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Schoof, M, Lawrence, R, Boone, M, Frost, A, Walter, P. | | Deposit date: | 2022-01-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A point mutation in the nucleotide exchange factor eIF2B constitutively activates the integrated stress response by allosteric modulation.
Elife, 11, 2022
|
|
7FAP
 
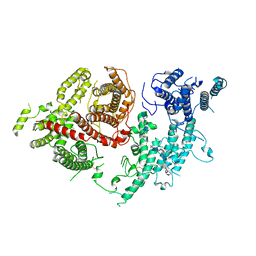 | | Structure of VAR2CSA-CSA 3D7 | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Wang, Z. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | Authors: | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
7CWS
 
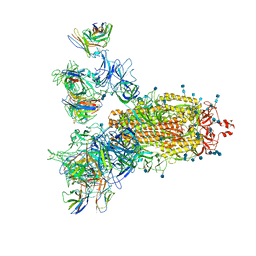 | | SARS-CoV-2 Spike Proteins Trimer in Complex with FC05 and H014 Fabs Cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of FC05 Fab, ... | | Authors: | Wang, L, Wang, X. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
3TG5
 
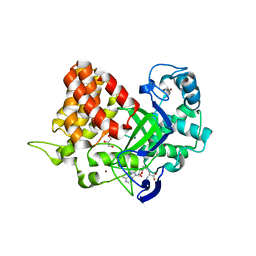 | | Structure of SMYD2 in complex with p53 and SAH | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
3CZO
 
 | |
5UDZ
 
 | |
3TG4
 
 | | Structure of SMYD2 in complex with SAM | | Descriptor: | GLYCEROL, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
3P12
 
 | | Crystal Structure of D-ribose Pyranase Sa240 | | Descriptor: | D-ribose pyranase, GLYCEROL | | Authors: | Wu, M, Wang, L, Zang, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus
J.Struct.Biol., 174, 2011
|
|
3P13
 
 | |
7K5I
 
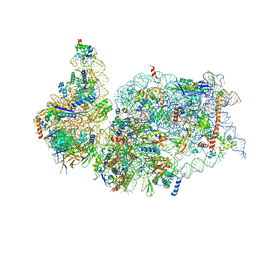 | | SARS-COV-2 nsp1 in complex with human 40S ribosome | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Wang, L, Shi, M, Wu, H. | | Deposit date: | 2020-09-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism.
Biorxiv, 2020
|
|
7L70
 
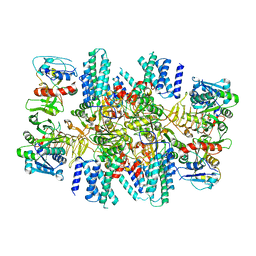 | | The eukaryotic translation initiation factor 2B from Homo sapiens in its apo form | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Schoof, M, Lawrence, R, Boone, M, Frost, A, Walter, P. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | eIF2B conformation and assembly state regulates the integrated stress response.
Elife, 10, 2021
|
|
2PVX
 
 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | Descriptor: | Rubredoxin, ZINC ION | | Authors: | Wang, L, LeMaster, D.M, Hernandez, G, Li, H. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin
Bmc Struct.Biol., 7, 2007
|
|
8JHU
 
 | | Legionella effector protein SidI | | Descriptor: | Legionella pneumophila effector protein SidI | | Authors: | Wang, L, Subramanian, A, Mukherjee, S, Walter, P. | | Deposit date: | 2023-05-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A Legionella toxin exhibits tRNA mimicry and glycosyl transferase activity to target the translation machinery and trigger a ribotoxic stress response.
Nat.Cell Biol., 25, 2023
|
|
7ENN
 
 | | The structure of ALC1 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromodomain-helicase-DNA-binding protein 1-like, ... | | Authors: | Chen, Z.C, Chen, K.J, Wang, L. | | Deposit date: | 2021-04-18 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
2IBJ
 
 | | Structure of House Fly Cytochrome B5 | | Descriptor: | Cytochrome b5, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L. | | Deposit date: | 2006-09-11 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparison of cytochromes b(5) from insects and vertebrates.
Proteins, 67, 2007
|
|
2I89
 
 | | Structure of septuple mutant of Rat Outer Mitochondrial Membrane Cytochrome B5 | | Descriptor: | Cytochrome b5 type B, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L, Sun, N. | | Deposit date: | 2006-09-01 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A histidine/tryptophan pi-stacking interaction stabilizes the heme-independent folding core of microsomal apocytochrome b5 relative to that of mitochondrial apocytochrome b5.
Biochemistry, 45, 2006
|
|
4V9D
 
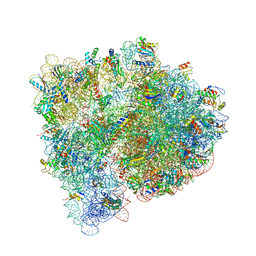 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
3ZPK
 
 | | Atomic-resolution structure of a quadruplet cross-beta amyloid fibril. | | Descriptor: | TRANSTHYRETIN | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S.A, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY, SOLID-STATE NMR | | Cite: | Atomic Structure and Hierarchical Assembly of a Cross-Beta Amyloid Fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3BIY
 
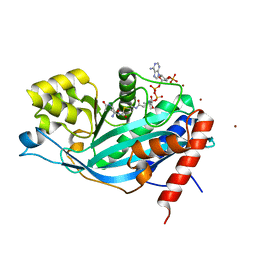 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
1QBQ
 
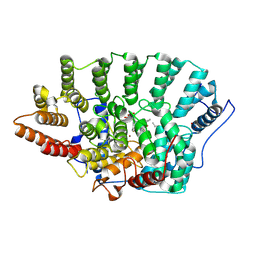 | | STRUCTURE OF RAT FARNESYL PROTEIN TRANSFERASE COMPLEXED WITH A CVIM PEPTIDE AND ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID. | | Descriptor: | ACETATE ION, ACETYL-CYS-VAL-ILE-SELENOMET-COOH PEPTIDE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, ... | | Authors: | Strickland, C.L, Windsor, W.T, Syto, R, Wang, L, Bond, R, Wu, Z, Schwartz, J, Le, H.V, Beese, L.S, Weber, P.C. | | Deposit date: | 1999-04-27 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue
Biochemistry, 37, 1998
|
|
2QWQ
 
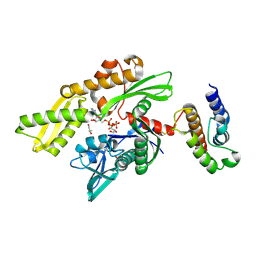 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-394aa)R171C and bovine Auxilin (810-910aa)D876C in the AMPPNP hydrolyzed form | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
