7RKZ
 
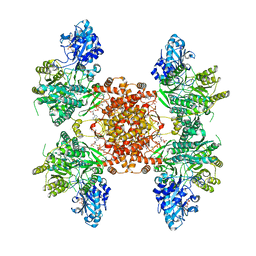 | | Structure of ACLY D1026A-substrates-asym-int | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-22 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7RMP
 
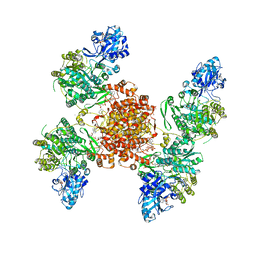 | | Structure of ACLY D1026A - substrates-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
6UUW
 
 | | Structure of human ATP citrate lyase E599Q mutant in complex with Mg2+, citrate, ATP and CoA | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-11-01 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UI9
 
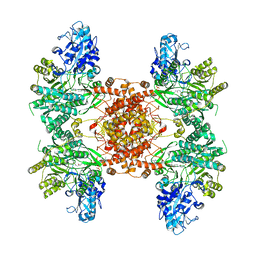 | |
6UV5
 
 | |
6VP9
 
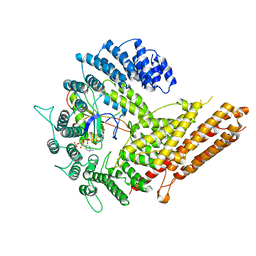 | | Cryo-EM structure of human NatB complex | | Descriptor: | CARBOXYMETHYL COENZYME *A, MDVFM peptide, N-alpha-acetyltransferase 20, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for N-terminal alpha-synuclein acetylation by human NatB.
Elife, 9, 2020
|
|
2X0I
 
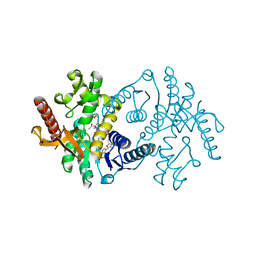 | | 2.9 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.-K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
3RJ5
 
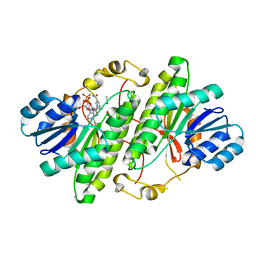 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
2X0J
 
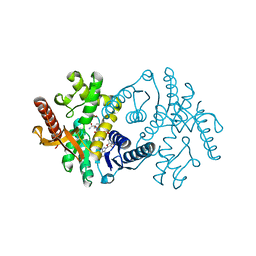 | | 2.8 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH ETHENO-NAD | | Descriptor: | ETHENO-NAD, MALATE DEHYDROGENASE, SULFATE ION | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
3RJ9
 
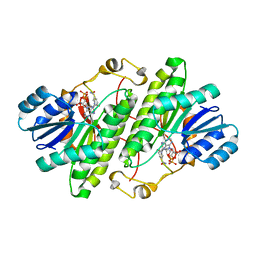 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
1W19
 
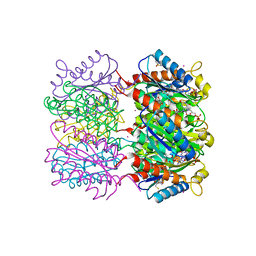 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1O7A
 
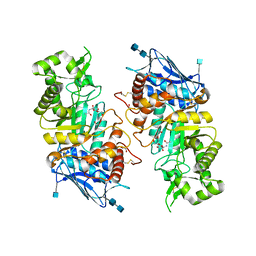 | | Human beta-Hexosaminidase B | | Descriptor: | 1,2-ETHANEDIOL, 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maier, T, Strater, N, Schuette, C, Klingenstein, R, Sandhoff, K, Saenger, W. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The X-Ray Crystal Structure of Human Beta-Hexosaminidase B Provides New Insights Into Sandhoff Disease
J.Mol.Biol., 328, 2003
|
|
1W29
 
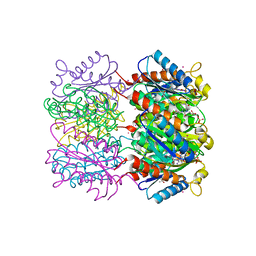 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)butane 1-phosphate | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 4-{2,6,8-TRIOXO-9-[(2R,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, 4-{2,6,8-TRIOXO-9-[(2S,3R,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-07-01 | | Release date: | 2005-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
4DAW
 
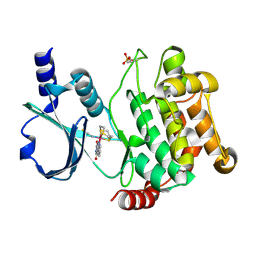 | | Crystal structure of PAK1 kinase domain with the ruthenium phthalimide complex | | Descriptor: | Serine/threonine-protein kinase PAK 1, [1,3-dioxo-6-(pyridin-2-yl-kappaN)-2,3-dihydro-1H-isoindol-5-yl-kappaC~5~][(thioxomethylidene)azanido-kappaN](1,4,7-trithionane-kappa~3~S~1~,S~4~,S~7~)ruthenium | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The art of filling protein pockets efficiently with octahedral metal complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1YK4
 
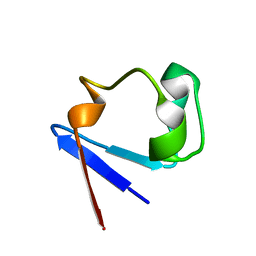 | | Ultra-high resolution structure of Pyrococcus abyssi rubredoxin W4L/R5S | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Bonisch, H, Schmidt, C.L, Bianco, P, Ladenstein, R. | | Deposit date: | 2005-01-17 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.69 Å) | | Cite: | Ultrahigh-resolution study on Pyrococcus abyssi rubredoxin. I. 0.69 A X-ray structure of mutant W4L/R5S.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3LBX
 
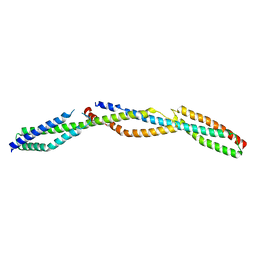 | | Crystal Structure of the Erythrocyte Spectrin Tetramerization Domain Complex | | Descriptor: | Spectrin alpha chain, erythrocyte, Spectrin beta chain | | Authors: | Ipsaro, J.J, Harper, S.L, Messick, T.E, Marmorstein, R, Mondragon, A, Speicher, D.W. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional interpretation of the erythrocyte spectrin tetramerization domain complex.
Blood, 115, 2010
|
|
4K2J
 
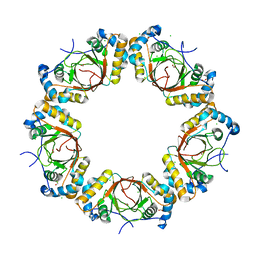 | |
1YK5
 
 | | Pyrococcus abyssi rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Bonisch, H, Schmidt, C.L, Bianco, P, Ladenstein, R. | | Deposit date: | 2005-01-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ultrahigh-resolution study on Pyrococcus abyssi rubredoxin. I. 0.69 A X-ray structure of mutant W4L/R5S.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3CSF
 
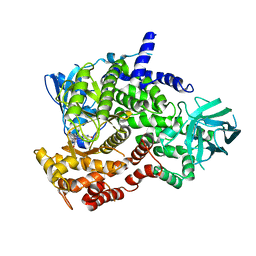 | |
2B9D
 
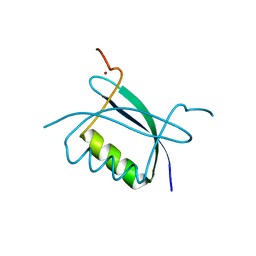 | | Crystal Structure of HPV E7 CR3 domain | | Descriptor: | E7 protein, ZINC ION | | Authors: | Liu, X, Clements, A, Zhao, K, Marmorstein, R. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor.
J.Biol.Chem., 281, 2006
|
|
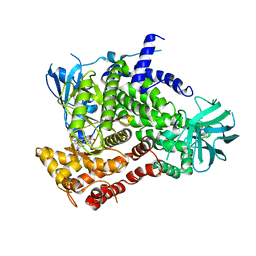
3CST
 
 | |
3BIY
 
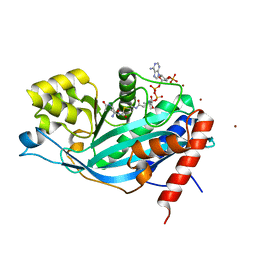 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
3DM7
 
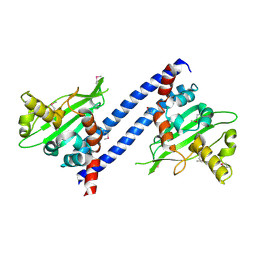 | |
2C49
 
 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | ADENOSINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-17 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C4E
 
 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | MAGNESIUM ION, SUGAR KINASE MJ0406 | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
