8XLQ
 
 | |
5XFF
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550L) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
8JI4
 
 | | Crystal structure of AetD in complex with 5-bromo-L-tryptophan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-bromo-L-tryptophan, AetD, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI5
 
 | | Crystal structure of AetD in complex with 5-bromo-L-tryptophan and two Fe2+ | | Descriptor: | 5-bromo-L-tryptophan, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI3
 
 | | Crystal structure of AetD in complex with 5,7-dibromo-L-tryptophan and two Fe2+ | | Descriptor: | (2S)-2-azanyl-3-[5,7-bis(bromanyl)-1H-indol-3-yl]propanoic acid, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI6
 
 | | Crystal structure of AetD in complex with L-tryptophan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI2
 
 | | Crystal structure of AetD in complex with 5,7-dibromo-L-tryptophan | | Descriptor: | (2S)-2-azanyl-3-[5,7-bis(bromanyl)-1H-indol-3-yl]propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AetD, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JZ3
 
 | | Crystal structure of AetF in complex with FAD and L-tryptophan | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JZ5
 
 | | Crystal structure of AetF in complex with FAD and NADP+ at 1.86 angstrom | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JZ2
 
 | | Crystal structure of AetF in complex with FAD | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JZ4
 
 | | Crystal structure of AetF in complex with FAD and 5-bromo-L-tryptophan | | Descriptor: | 5-bromo-L-tryptophan, AetF, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
6WGN
 
 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | Descriptor: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
8JI7
 
 | | Crystal structure of AetD in complex with L-tryptophan and two Fe2+ | | Descriptor: | AetD, FE (II) ION, NICKEL (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
3LD8
 
 | | Structure of JMJD6 and Fab Fragments | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, GLYCEROL, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LDB
 
 | | Structure of JMJD6 complexd with ALPHA-KETOGLUTARATE and Fab Fragment. | | Descriptor: | 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8DOT
 
 | |
4P57
 
 | | MHC TCR peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Clayton, G.M, Crawford, F, Kappler, J.W. | | Deposit date: | 2014-03-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
4P4R
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergic Hypersensitivity and Autoimmunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DP alpha 1 chain, ... | | Authors: | Clayton, G.M, Crawford, F, Kappler, J.W. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
4P4K
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between allergic hypersensitivity and auto immunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM, HLA class II histocompatibility antigen, ... | | Authors: | Clayton, G.M, Crawford, F, Kappler, J.W. | | Deposit date: | 2014-03-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
4KEG
 
 | | Crystal Structure of MBP Fused Human SPLUNC1 | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic/Palate lung and nasal epithelium clone fusion protein, octyl beta-D-glucopyranoside | | Authors: | Ning, F, Wang, C, Niu, L, Chu, H.W, Zhang, G. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Ligands of the SPLUNC1 Protein
To be Published
|
|
1OQD
 
 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1OQE
 
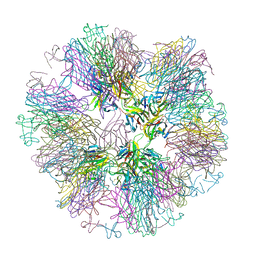 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
7F3M
 
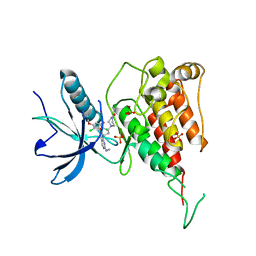 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
8HBM
 
 | |
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
