6SZ7
 
 | | Crystal structure of YTHDC1 with fragment 5 (DHU_DC1_066) | | Descriptor: | 5,6,7,8-tetrahydro-4~{a}~{H}-quinazoline-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3D04
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with sakuranetin | | Descriptor: | (2S)-5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-2,3-dihydro-4H-chromen-4-one, (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, ... | | Authors: | Zhang, L, Kong, Y, Wu, D, Shen, X, Jiang, H. | | Deposit date: | 2008-05-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
2NCY
 
 | | Solution structure of pseudin-2 analog (Ps-P) | | Descriptor: | Pseudin-2 | | Authors: | Jeon, D, Kim, J, Shin, A, Kim, Y. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimum Balance between the Cationicity and Structural Component for Bacterial Cell Selectivity and Anti-inflammatory activities of Pseudin-2 and its Analogs
To be Published
|
|
6SZR
 
 | | Crystal structure of YTHDC1 with fragment 9 (DHU_DC1_107) | | Descriptor: | 6-[[methyl-(phenylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
1FHT
 
 | | RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1996-02-21 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding.
J.Mol.Biol., 257, 1996
|
|
3JRA
 
 | |
6SZX
 
 | | Crystal structure of YTHDC1 with fragment 11 (DHU_DC1_128) | | Descriptor: | 6-[[cyclopropyl-(phenylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T06
 
 | | Crystal structure of YTHDC1 with fragment 19 (DHU_DC1_045) | | Descriptor: | 3-imidazolidin-2-yl-2~{H}-indazole, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0C
 
 | | Crystal structure of YTHDC1 with fragment 26 (DHU_DC1_198) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-2~{H}-indazole-3-carboxamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3D2P
 
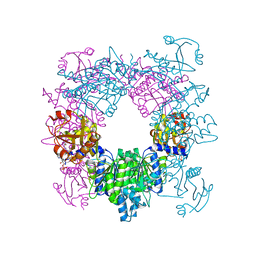 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-arginine | | Descriptor: | ARGININE, COENZYME A, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
3JUZ
 
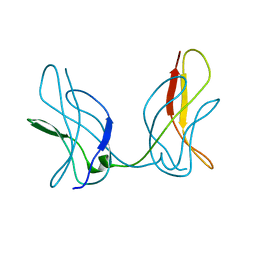 | |
4L54
 
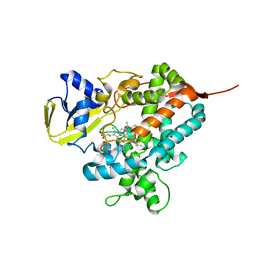 | | Structure of cytochrome P450 OleT, ligand-free | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Terminal olefin-forming fatty acid decarboxylase | | Authors: | Leys, D. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Properties of the Alkene Producing Cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 Bacterium.
J.Biol.Chem., 289, 2014
|
|
8AOL
 
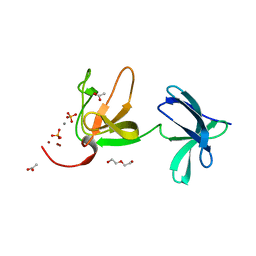 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain III (aa 363-499) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sagmeister, T, Damisch, E, Eder, M, Dordic, A, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6SPD
 
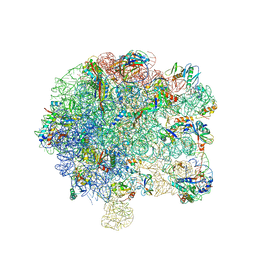 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SS5
 
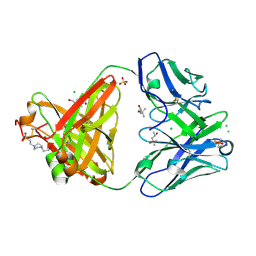 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3JWE
 
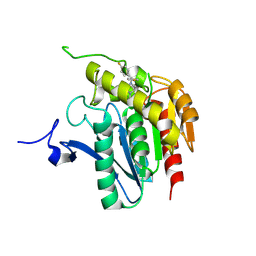 | | Crystal structure of human mono-glyceride lipase in complex with SAR629 | | Descriptor: | 1-[bis(4-fluorophenyl)methyl]-4-(1H-1,2,4-triazol-1-ylcarbonyl)piperazine, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
3JWP
 
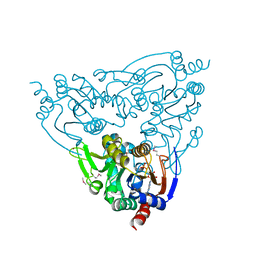 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
6SY0
 
 | |
6SSH
 
 | | Structure of the TSC2 GAP domain | | Descriptor: | 1,2-ETHANEDIOL, GTPase activator-like protein | | Authors: | Hansmann, P, Kiontke, S, Kummel, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the TSC2 GAP Domain: Mechanistic Insight into Catalysis and Pathogenic Mutations.
Structure, 28, 2020
|
|
1FPN
 
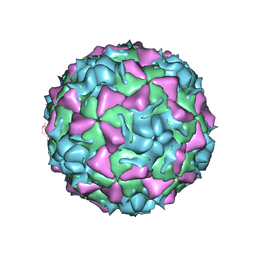 | | HUMAN RHINOVIRUS SEROTYPE 2 (HRV2) | | Descriptor: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | Authors: | Verdaguer, N, Blaas, D, Fita, I. | | Deposit date: | 2000-08-31 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human rhinovirus serotype 2 (HRV2).
J.Mol.Biol., 300, 2000
|
|
1FQF
 
 | |
3JYH
 
 | | Human dipeptidyl peptidase DPP7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl-peptidase 2, ... | | Authors: | Dobrovetsky, E, Dong, A, Seitova, A, Crombett, L, Paganon, S, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Hassel, A, Shewchuk, L, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Human dipeptidyl peptidase DPP7
To be Published
|
|
3ZD7
 
 | | Snapshot 3 of RIG-I scanning on RNA duplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-11-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Defining the Functional Determinants for RNA Surveillance by Rig-I.
Embo Rep., 14, 2013
|
|
6T0O
 
 | | Crystal structure of YTHDC1 with fragment 14 (ACA_DC1_004) | | Descriptor: | 2-methyl-3~{H}-pyrido[3,4-d]pyrimidin-4-one, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3J1V
 
 | |
