8H6P
 
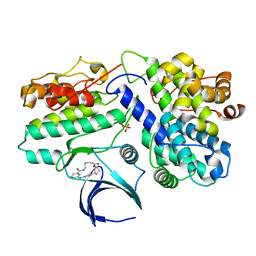 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
7F3H
 
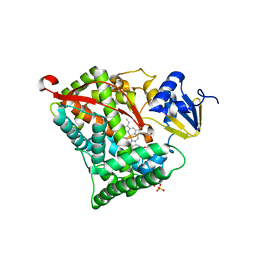 | | Crystal structure of cytochrome P450DA heme domain | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SERINE, ... | | Authors: | Wan, N.W. | | Deposit date: | 2021-06-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution of cytochrome P450DA hydroxylase activity for stereoselective biohydroxylation
Catalysis Science And Technology, 12, 2022
|
|
7X4B
 
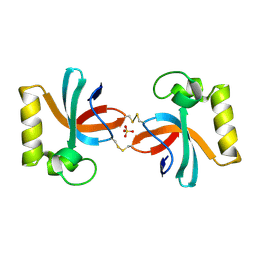 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7XCZ
 
 | |
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | Authors: | Liu, Z, Lui, S, Gao, Y. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDA
 
 | |
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
6L7D
 
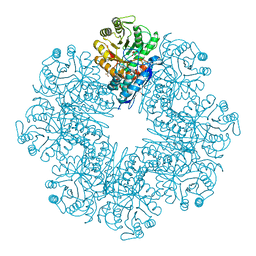 | | Mycobacterium tuberculosis enolase mutant - S42A | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, ACETATE ION, ... | | Authors: | Ahmad, M, Jha, B, Tiwari, S, Dwivedy, A, Biswal, B.K. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
7X31
 
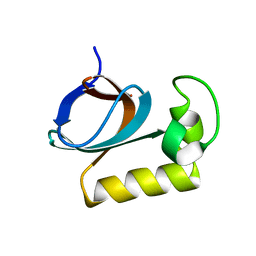 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
5UOJ
 
 | |
5UQ9
 
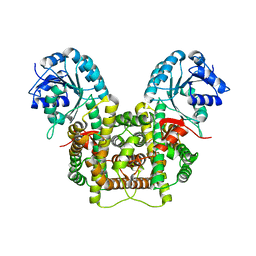 | | Crystal structure of 6-phosphogluconate dehydrogenase with ((4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl)methyl dihydrogen phosphate | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating, [(4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl]methyl dihydrogen phosphate | | Authors: | Leonard, P.G. | | Deposit date: | 2017-02-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation.
Cell Rep, 26, 2019
|
|
4V4B
 
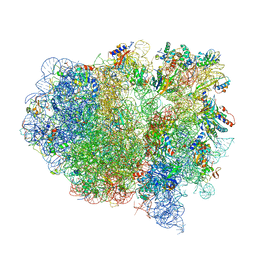 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
7CYU
 
 | | Crystal structure of human BAF57 HMG domain | | Descriptor: | SULFATE ION, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 | | Authors: | Heo, Y, Yun, J.H, Park, J.H, Lee, W. | | Deposit date: | 2020-09-04 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the HMG domain of human BAF57 and its interaction with four-way junction DNA.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7E8I
 
 | | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (145-MER), Histone H2A, ... | | Authors: | Dai, Y, Dai, L, Zhou, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin.
Mol.Cell, 81, 2021
|
|
3OUM
 
 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2010-09-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
3OUL
 
 | |
6DQ4
 
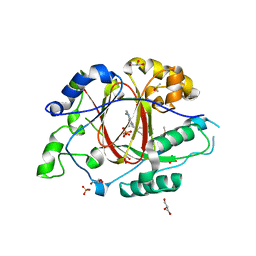 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6NZQ
 
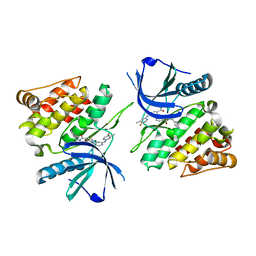 | |
4U02
 
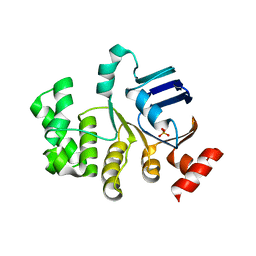 | | Crystal structure of apo-TTHA1159 | | Descriptor: | Amino acid ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | Karthiga Devi, S, Chichili, V.P.R, Velmurugan, D, Sivaraman, J. | | Deposit date: | 2014-07-11 | | Release date: | 2015-05-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus
J.Struct.Biol., 190, 2015
|
|
6DQ9
 
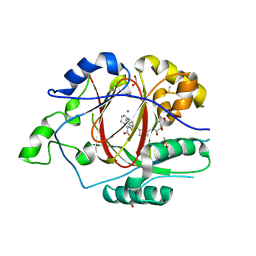 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
4OWI
 
 | | peptide structure | | Descriptor: | p53LZ2 | | Authors: | Lee, J.-H. | | Deposit date: | 2014-02-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Protein grafting of p53TAD onto a leucine zipper scaffold generates a potent HDM dual inhibitor.
Nat Commun, 5, 2014
|
|
