7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
8CMF
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 nsp3 epitope (orf1ab)1350-1364 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CMI
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Omicron (BA.1) Spike peptide S761-775 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CME
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Membrane peptide M176-190 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CMD
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Spike peptide S761-775 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CMH
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Omicron (BA.1) Spike peptide S486-505 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CMC
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Spike peptide S511-530 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CMG
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 nsp14 peptide (orf1ab)6420-6434 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CMB
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Spike peptide S486-505 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
9EOU
 
 | |
6IZH
 
 | |
8HGV
 
 | |
8HGW
 
 | | Crystal structure of MehpH in complex with MBP | | Descriptor: | 1-BUTANOL, Monoalkyl phthalate hydrolase, PHTHALIC ACID | | Authors: | Zhang, Z.M, Wang, Y.J, Chen, Y.B. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.80001163 Å) | | Cite: | Molecular insights into the catalytic mechanism of plasticizer degradation by a monoalkyl phthalate hydrolase.
Commun Chem, 6, 2023
|
|
8X61
 
 | | Cryo-EM structure of ATP-bound FtsE(E163Q)X | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX | | Authors: | Zhang, Z.Y, Chen, Y.T. | | Deposit date: | 2023-11-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
6KHY
 
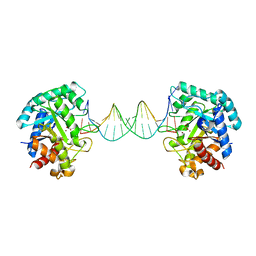 | | The crystal structure of AsfvAP:AG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
2PKG
 
 | |
6IMK
 
 | | The crystal structure of AsfvLIG:CG complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*G)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
6IMJ
 
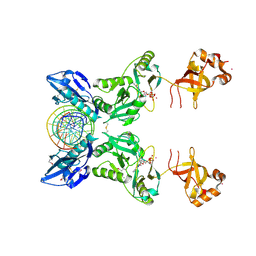 | | The crystal structure of Se-AsfvLIG:DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
6IMN
 
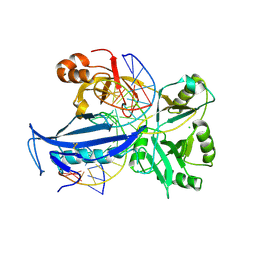 | | The crystal structure of AsfvLIG:CT2 complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
6IML
 
 | | The crystal structure of AsfvLIG:CT1 complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
8X2O
 
 | | RIPK2 in complex with K252 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[(1~{R})-4-[4-[(6-fluoranyl-1,3-benzothiazol-5-yl)amino]thieno[2,3-d]pyrimidin-6-yl]cyclohex-3-en-1-yl]cyclopropanecarboxamide | | Authors: | Yang, J.H, Yang, J.H. | | Deposit date: | 2023-11-10 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | CMD-OPT model enables the discovery of a potent and selective RIPK2 inhibitor as preclinical candidate for the treatment of acute liver injury
Acta Pharm Sin B, 2025
|
|
7V61
 
 | | ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3E8, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker.
Signal Transduct Target Ther, 6, 2021
|
|
7Y1F
 
 | | Cryo-EM structure of human k-opioid receptor-Gi complex | | Descriptor: | Dynorphin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, B.O, Xu, F.E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of human kappa-opioid receptor-Gi complex bound to an endogenous agonist dynorphin A.
Protein Cell, 14, 2023
|
|
7KHF
 
 | |
