2EFW
 
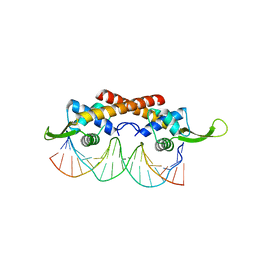 | | Crystal structure of the RTP:nRB complex from Bacillus subtilis | | Descriptor: | DNA (5'-D(*DCP*DT*DAP*DTP*DGP*DTP*DAP*DCP*DCP*DAP*DAP*DAP*DTP*DGP*DTP*DTP*DCP*DAP*DGP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DCP*DTP*DGP*DAP*DAP*DCP*DAP*DTP*DTP*DTP*DGP*DGP*DTP*DAP*DCP*DAP*DTP*DAP*DG)-3'), Replication termination protein | | Authors: | Vivian, J.P, Porter, C.J, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2007-02-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA
J.Mol.Biol., 370, 2007
|
|
2XLK
 
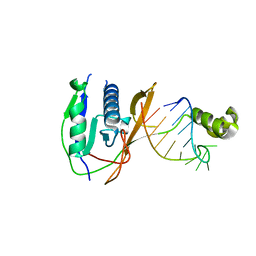 | | Crystal structure of the Csy4-crRNA complex, orthorhombic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2XTE
 
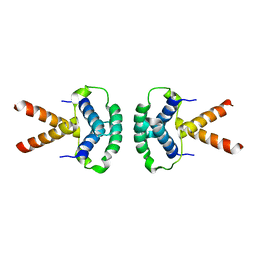 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7ZR2
 
 | | Crystal structure of a chimeric protein mimic of SARS-CoV-2 Spike HR1 in complex with HR2 | | Descriptor: | Spike protein S2', Spike protein S2',Chimeric protein mimic of SARS-CoV-2 Spike HR1 | | Authors: | Camara-Artigas, A, Gavira, J.A, Cano-Munoz, M, Polo-Megias, D, Conejero-Lara, F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel chimeric proteins mimicking SARS-CoV-2 spike epitopes with broad inhibitory activity.
Int.J.Biol.Macromol., 222, 2022
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2COQ
 
 | |
8A4L
 
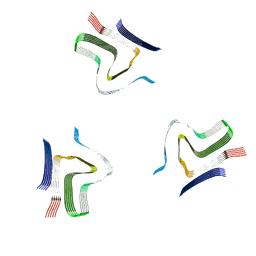 | | Lipidic alpha-synuclein fibril - polymorph L2A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-06-12 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
2CJM
 
 | | Mechanism of CDK inhibition by active site phosphorylation: CDK2 Y15p T160p in complex with cyclin A structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Welburn, J.P.I, Tucker, J, Johnson, T, Lindert, L, Morgan, M, Willis, A, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How Tyrosine 15 Phosphorylation Inhibits the Activity of Cyclin-Dependent Kinase 2-Cyclin A.
J.Biol.Chem., 282, 2007
|
|
3NN8
 
 | |
8A56
 
 | | Coenzyme A-persulfide reductase (CoAPR) from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Coenzyme A-persulfide reductase, ... | | Authors: | Costa, S.S, Walsh, B.J, Giedroc, D.P, Brito, J.A. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Metabolic and Structural Insights into Hydrogen Sulfide Mis-Regulation in Enterococcus faecalis.
Antioxidants, 11, 2022
|
|
8A7N
 
 | | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125) | | Descriptor: | N-(3-azanylpropyl)-3-[[(3S)-1-(2-fluorophenyl)-2-oxidanylidene-pyrrolidin-3-yl]amino]-N-methyl-benzamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-06-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125)
To Be Published
|
|
8A10
 
 | | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-020-049-143 | | Descriptor: | T-box transcription factor T, UNKNOWN LIGAND | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-020-049-143
To Be Published
|
|
7ZMS
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G4-043 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G4-043, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
8B58
 
 | |
2AYR
 
 | | A SERM Designed for the Treatment of Uterine Leiomyoma with Unique Tissue Specificity for Uterus and Ovaries in Rats | | Descriptor: | 6-(4-METHYLSULFONYL-PHENYL)-5-[4-(2-PIPERIDIN-1-YLETHOXY)PHENOXY]NAPHTHALEN-2-OL, Estrogen receptor | | Authors: | Hummel, C.W, Geiser, A.G, Bryant, H.U, Cohen, I.R, Dally, R.D, Fong, K.C, Frank, S.A, Hinklin, R, Jones, S.A, Lewis, G, McCann, D.J, Shepherd, T.A, Tian, H, Rudman, D.G, Wallace, O.B, Wang, Y, Dodge, J.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A selective estrogen receptor modulator designed for the treatment of uterine leiomyoma with unique tissue specificity for uterus and ovaries in rats
J.Med.Chem., 48, 2005
|
|
2B4X
 
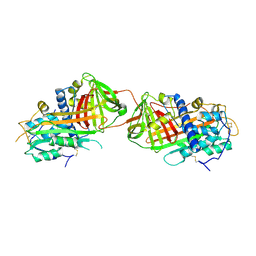 | |
2YOF
 
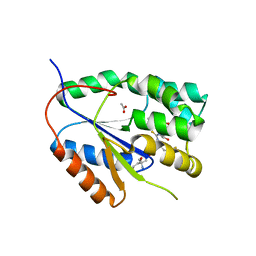 | | Plasmodium falciparum thymidylate kinase in complex with a (thio)urea- beta-deoxythymidine inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[[(2R,3S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methyl]thiourea, ACETATE ION, THYMIDYLATE KINASE, ... | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
2YBD
 
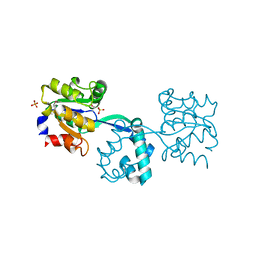 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
2YDJ
 
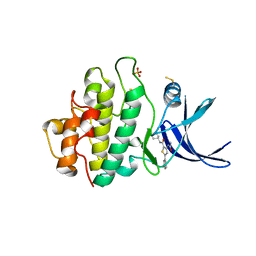 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, PHOSPHATE ION, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-01-25 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
372D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
397D
 
 | |
2BAY
 
 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
2AV8
 
 | | Y122F MUTANT OF RIBONUCLEOTIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | FE (II) ION, MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
3Q8M
 
 | | Crystal Structure of Human Flap Endonuclease FEN1 (D181A) in complex with substrate 5'-flap DNA and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
2AWM
 
 | | GFP R96A chromophore maturation recovery mutant R96A Q183R | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
