8C5L
 
 | |
6GIP
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 2, 5-dimethyl core. | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dimethyl-6-quinolin-4-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mathea, S, Chen, Z, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
4AG5
 
 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8WB1
 
 | | Crystal Structure of small molecule KN2H covalently bound to K-Ras(G12D) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S},5~{R})-3-[7-(8-ethynyl-7-fluoranyl-3-oxidanyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]ethanone, GTPase KRas, ... | | Authors: | Zhou, L, Zhang, R, Shuai, T.B. | | Deposit date: | 2023-09-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of small molecule KN2H covalently bound to K-Ras(G12D)
To Be Published
|
|
1OF1
 
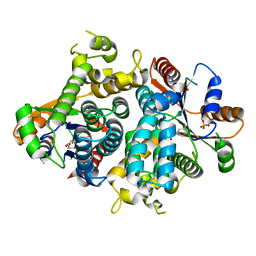 | | KINETICS AND CRYSTAL STRUCTURE OF THE HERPES SIMPLEX VIRUS TYPE 1 THYMIDINE KINASE INTERACTING WITH (SOUTH)-METHANOCARBA-THYMIDINE | | Descriptor: | (SOUTH)-METHANOCARBA-THYMIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Claus, M.T, Schelling, P, Folkers, G, Marquez, V.E, Scapozza, L, Schulz, G.E. | | Deposit date: | 2003-04-03 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and Structural Characterization of (South)-Methanocarbathymidine that Specifically Inhibits Growth of Herpes Simplex Virus Type 1 Thymidine Kinase-Transduced Osteosarcoma Cells
J.Biol.Chem., 279, 2004
|
|
1CM0
 
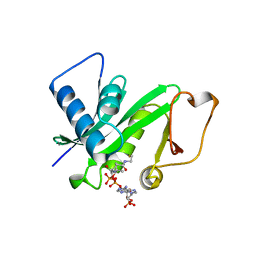 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
5N62
 
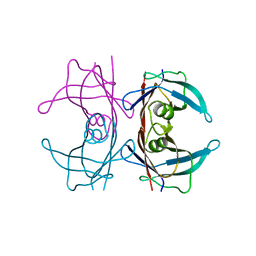 | | Human TTR crystals soaked in manganese chloride. | | Descriptor: | MANGANESE (II) ION, Transthyretin | | Authors: | Ciccone, L, Savko, M, Shepard, W, Stura, E.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Copper mediated amyloid-beta binding to Transthyretin.
Sci Rep, 8, 2018
|
|
8WBZ
 
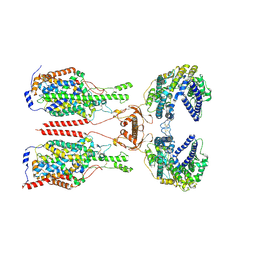 | | Cryo-EM structure of ACE2-B0AT1 complex with JX225 | | Descriptor: | 2-(4-bromanyl-3-methyl-phenoxy)-~{N}-propyl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R, Hu, Z, Dai, L. | | Deposit date: | 2023-09-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of inhibition of the amino acid transporter B 0 AT1 (SLC6A19).
Nat Commun, 15, 2024
|
|
5N6I
 
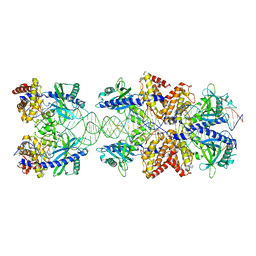 | | Crystal structure of mouse cGAS in complex with 39 bp DNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA (36-MER), DNA (37-MER), ... | | Authors: | Andreeva, L, Kostrewa, D, Hopfner, K.-P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders.
Nature, 549, 2017
|
|
5YXU
 
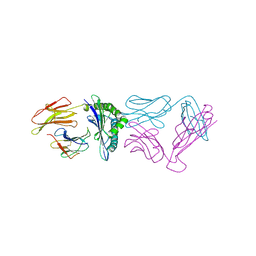 | |
6GLE
 
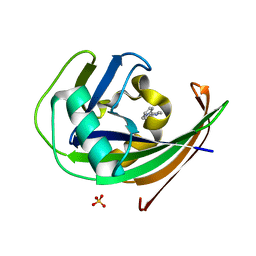 | | Crystal structure of hMTH1 in complex with TH scaffold 1 in the presence of acetate | | Descriptor: | 4-phenylpyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | hMTH1 in complex with TH scaffold 1
To Be Published
|
|
6GLK
 
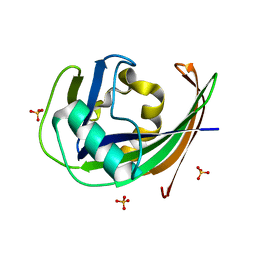 | |
6GLR
 
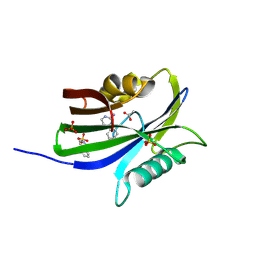 | | Crystal structure of hMTH1 N33G in complex with TH scaffold 1 in the presence of acetate | | Descriptor: | 4-phenylpyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | hMTH1 N33G in complex with TH scaffold 1
To Be Published
|
|
1NNI
 
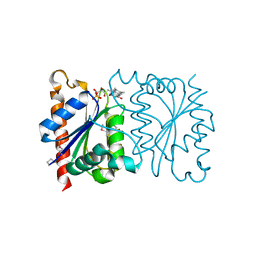 | | Azobenzene Reductase from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein yhda | | Authors: | Cuff, M.E, Kim, Y, Maj, L, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-13 | | Release date: | 2003-07-29 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azobenzene Reductase from Bacillus subtilis
To be Published, 2003
|
|
4ANG
 
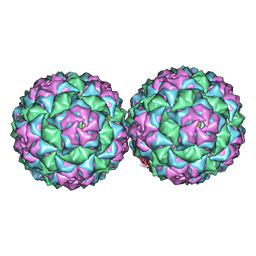 | | Small RNA phage PRR1 in complex with an RNA operator fragment | | Descriptor: | 5'-R(*CP*CP*AP*UP*AP*AP*GP*GP*AP*GP*CP*UP*AP*CP *CP*UP*AP*UP*GP*GP)-3', CALCIUM ION, COAT PROTEIN | | Authors: | Persson, M, Tars, K, Liljas, L. | | Deposit date: | 2012-03-16 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Prr1 Coat Protein Binding to its RNA Translational Operator
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4A0S
 
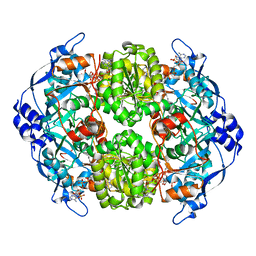 | | STRUCTURE OF THE 2-OCTENOYL-COA CARBOXYLASE REDUCTASE CINF IN COMPLEX WITH NADP AND 2-OCTENOYL-COA | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A, OCTENOYL-COA REDUCTASE/CARBOXYLASE | | Authors: | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | Deposit date: | 2011-09-12 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual carbon fixation gives rise to diverse polyketide extender units.
Nat. Chem. Biol., 8, 2011
|
|
4A31
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 6-{[2-(4-METHYLPIPERAZIN-1-YL)ETHYL]AMINO}-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)PYRIDINE-3-SULFONAMIDE, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
8SPA
 
 | | Structural insights into cellular control of the human CPEB3 prion, functionally regulated by a labile-amyloid-forming segment | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Flores, M.D, Sawaya, M.R, Boyer, D.R, Zink, S, Fioriti, L, Rodriguez, J.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a reversible amyloid fibril formed by the CPEB3 prion-like domain reveals a core sequence involved in translational regulation
To Be Published
|
|
1NLM
 
 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | Descriptor: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8C1J
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Protein arginine N-methyltransferase 2, RNA-binding protein Rsf1-like | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30
To Be Published
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5NCQ
 
 | | Structure of the (SR) Ca2+-ATPase bound to a Tetrahydrocarbazole and TNP-ATP | | Descriptor: | (1~{S})-~{N}-[(4-bromophenyl)methyl]-7-(trifluoromethyloxy)-2,3,4,9-tetrahydro-1~{H}-carbazol-1-amine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, ... | | Authors: | Bublitz, M, Kjellerup, L, O'Hanlon Cohrt, K, Gordon, S, Mortensen, A.L, Clausen, J.D, Pallin, D, Hansen, J.B, Brown, W.D, Fuglsang, A, Winther, A.-M.L. | | Deposit date: | 2017-03-06 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetrahydrocarbazoles are a novel class of potent P-type ATPase inhibitors with antifungal activity.
PLoS ONE, 13, 2018
|
|
8VJL
 
 | | SpoIVFB:pro-sigmaK complex | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Lauryl Maltose Neopentyl Glycol, RNA polymerase sigma factor, ... | | Authors: | Orlando, M.A, Pouillon, H.J.T, Mandal, S, Kroos, L, Orlando, B.J. | | Deposit date: | 2024-01-07 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Substrate engagement by the intramembrane metalloprotease SpoIVFB.
Nat Commun, 15, 2024
|
|
6GGV
 
 | | Structure of the arginine-bound form of truncated (residues 20-233) ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Smaldone, G, Berisio, R, Balasco, N, D'Auria, S, Vitagliano, L, Ruggiero, A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Domain swapping dissection in Thermotoga maritima arginine binding protein: How structural flexibility may compensate destabilization.
Biochim. Biophys. Acta, 1866, 2018
|
|
1CWX
 
 | | SOLUTION STRUCTURE OF THE HEPATITIS C VIRUS N-TERMINAL CAPSID PROTEIN 2-45 [C-HCV(2-45)] | | Descriptor: | HEPATITIS C VIRUS CAPSID PROTEIN | | Authors: | Ladaviere, L, Deleage, G, Montserret, R, Dalbon, P, Jolivet, M, Penin, F. | | Deposit date: | 1999-08-27 | | Release date: | 1999-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Immunodominant Antigenic Region of the Hepatitis C Virus Capsid Protein by NMR
To be Published
|
|
